FIGURE 1.
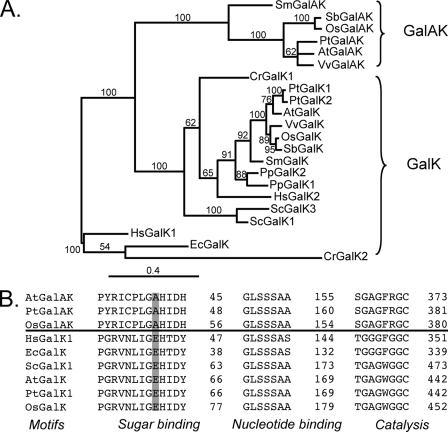
Sequence alignments and phylogenetic relationships of GalAK and GalK from different organisms. A, phylogenetic relationships of GalAK and GalK in different species. Protein sequences (see name and gene accession numbers below) were aligned and analyzed using Muscle 3.7 software (38) and the phylogenetic trees were created using MrBayes 3.1.2 software (39). Branch support values (more than 50%) are shown. The bar represents 0.4 protein substitutions per site. B, the conserved motifs of the putative GalAK sugar-binding, ATP-binding, and catalytic domains were aligned with GalK from different species. Note: the conserved AA Glu in the sugar binding motif of GalK is altered in GalAK to Ala. A. thaliana GalA-kinase (AtGalAK, At3g10700), Oryza sativa GalAK (OsGalAK, Os4g51880), Populus trichocarpa GalAK (PtGalAK, JGI:427630), Vitis vinifera GalAK (VvGalAK, GSVIVT00007137001), Sorghum bicolor GalAK (SbGalAK, Sb06g027910), Selaginella moellendorffii GalAK (SmGalAK, JGI:82393), A. thaliana Gal kinase (AtGalK, At3g06580), GalK from Saccharomyces cerevisiae (ScGalK1, 2AJ4 and ScGalK3, NP_010292), GalK from Homo Sapiens (HsGalK1, 1WUU and HsGalK2, 2A2D), E. coli (EcGalK, NP_308812), Chlamydomonas reinhardtii (CrGalK1, XP_001701527 and CrGalK2, XP_001691828), Physcomitrella patens GalK (PpGalK1, XP_001767042 and PpGalK2, XP_001756018), O. sativa (OsGalK, Os03g0832600), Populus trichocarpa (PtGalK1, XP_002314964 and PtGalK2, XP_002312315), V. vinifera (VvGalK, GSVIVT00033170001), S. bicolor (SbGalK, Sb01g002480), and S. moellendorffii (SmGalK, JGI:144964).