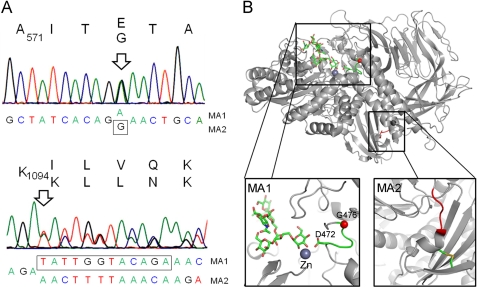
FIGURE 5.
Molecular basis of the Lec36 mutation. A, sequence of Golgi α-mannosidase II (MAN2A1) from HEK 293T Lec36 cells indicating the positions of the point mutation in MA1 encoding G574E and the in-frame deletion in MA2 resulting in the ΔILVQ deletion. B, mapping of the Lec36 mutations onto the crystal structure of D. melanogaster Golgi α-mannosidase II (Protein Data Bank ID code 3CZN) complexed with zinc and oligomannose-type substrate (56). The box on the left shows the area of the active site containing the residue analogous to the Lec36 G574E substitution encoded by MA1. The active site comprises a loop, depicted in green, containing both the ligand-binding Asp472 and the buried Gly476 (equivalent to Asp570 and Gly574, respectively, of the human enzyme). The Gly476 Cα carbon is depicted as a red sphere. The box on the right highlights in red the region analogous to the ΔILVQ (995QKLD998) deletion encoded by MA2. The molecular graphics images were generated using PyMOL (DeLano Scientific, San Carlos, CA).