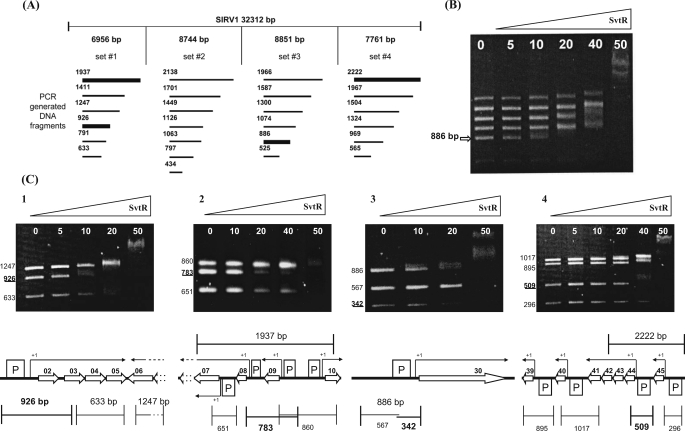
FIGURE 3.
Identification of the SvtR binding sites by the AG-EMSA approach. A, AG-EMSA strategy of the global protein-DNA interaction study for SIRV1. The SIRV1 genome was divided into four sets incubated separately with the SvtR protein in varying amounts (indicated in B and C in nanograms) and migrated on 1.2% agarose gels. The bold lines represent DNA fragments whose mobility changed specifically, i.e. at low concentrations of SvtR. B, typical results obtained with initial sets of DNA fragments (illustrated with set 3). The arrow indicates the position of the specifically responding fragment of 886 bp. C, AG-EMSA results that led to the identification of the SvtR targets and their positions on the genetic map of SIRV1. The strongest binding was observed in C3 with the gp30 promoter region. C1, a fragment of 926 bp was identified as specifically responding to SvtR in the first round of analysis. The scheme indicates this fragment's position that covered the promoter region for the operon gp02/03/04/05. C2, a 1937-bp fragment initially identified was divided into three subfragments. The 783-bp subfragment showed the strongest response to SvtR. This fragment covers the gp08 and gp09 promoters. because the gp09 promoter region was also included in a 860-bp fragment that did not specifically respond in the assay, it was concluded that the gp08 promoter region was involved in the specific binding of SvtR. C3, the initially identified 886-bp fragment was divided into two subfragments. In this analysis, the 342-bp fragment responded specifically. As indicated in the scheme, this fragment covers the gp30 promoter region. C4, a 2222-bp fragment covering two promoter regions was identified in the initial round of AG-EMSA. A 509-bp fragment was identified when subfragments were analyzed; its position covering the promoter region for the operon gp44-43-42-41 is indicated in the corresponding scheme.