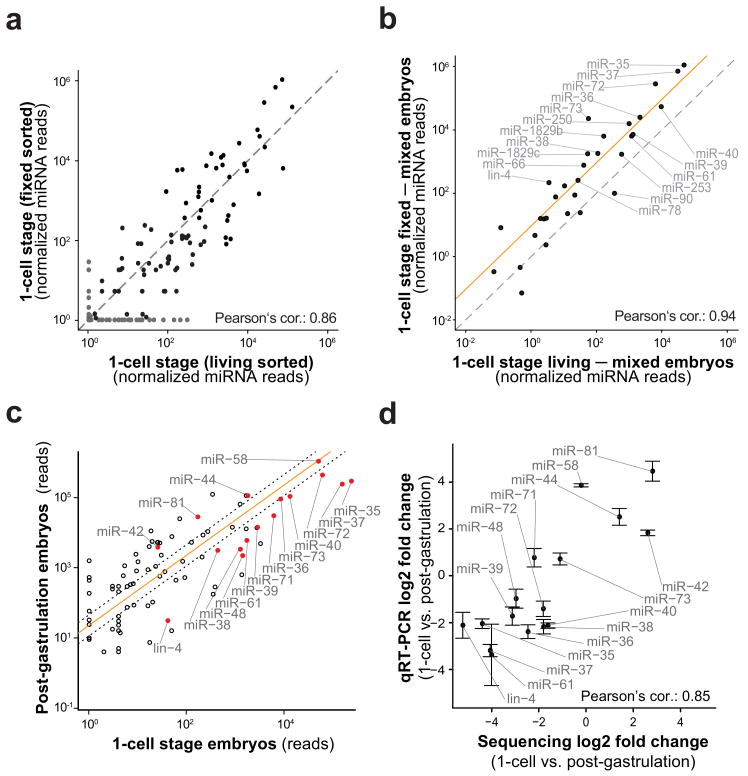
Figure 3.
FACS sorting of fixed one-cell embryos can be used to reveal miRNA expression dynamics.
(a) Scatter plot of miRNA expression quantified by normalized sequencing reads in the fixed resorted one-cell stage embryos (>98% purity) versus the live-sorted sample (∼70% one-cell embryos, ∼30% older embryos). The graph reveals a high correlation (Pearson 0.86) but considerable scatter and some miRNAs that seem exclusively expressed in only one of the samples (grey dots). Grey dashed line: main diagonal. (b) miRNA expression of live- and fixed-sorted one-cell stage embryos after in silico subtraction of miRNA expression in mixed embryos correlate almost perfectly (Pearson 0.94). The plot shows all miRNAs with higher expression in the one-cell embryos compared to the mixed embryos. Virtually all of these (linear regression, orange line) reside above the diagonal (grey dashed line) demonstrating the higher enrichment of one-cell specific miRNAs in the fixed-sorted sample. (c) Fold changes of miRNA expression between the one-cell stage embryos to post-gastrulation embryos as quantified by deep sequencing is shown in a double-logarithmic scatter plot. Orange line: linear regression. Dotted lines: 2 fold change. (d) qRT-PCRs for selected miRNAs (marked in red in (c)) were performed to assay miRNA fold changes on living, hand-picked embryos. Fold changes obtained by sequencing are plotted versus fold changes obtained by qRT-PCRs (Pearson 0.85). Coordinated expression fold changes are observed within the miRNA clusters miR-35 and miRNA-42 (Supplementary Fig. 3). Error bars s.d. (n=3).