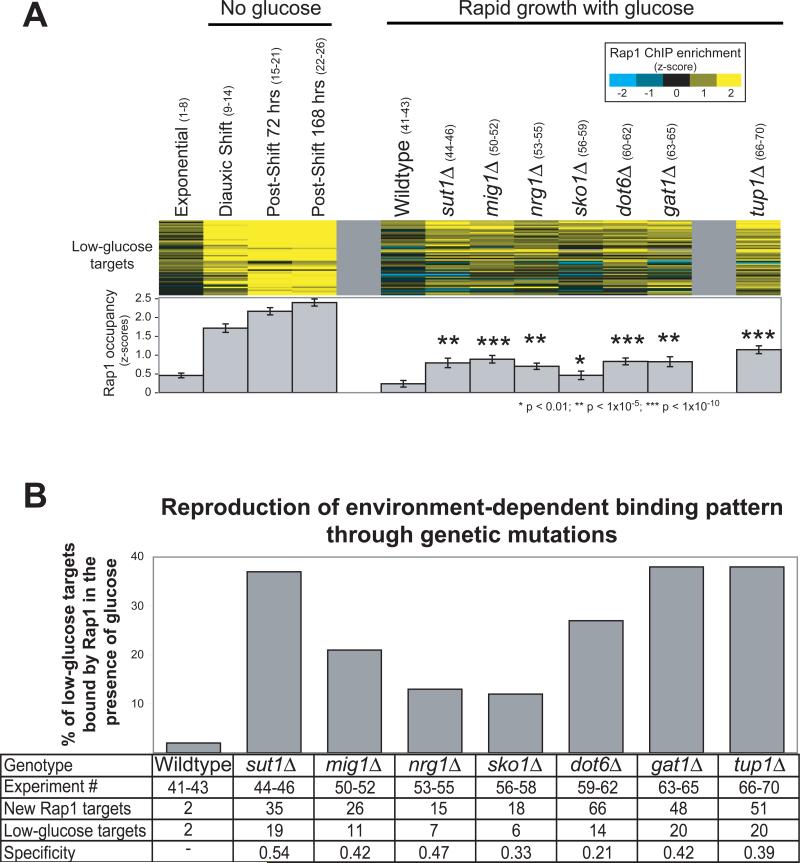
Figure 2. In the absence of Tup1 or proteins that recruit Tup1, Rap1 binds specifically and inappropriately to low-nutrient targets.
A) Rap1 occupancy determined by ChIP is shown at the 52 low-glucose targets in the indicated mutant strains (z-scores for enrichment, scalebar upper right). Experiments 1-26 (shown in parentheses, detailed in Table S1) measure Rap1 occupancy over a timecourse as glucose is depleted from the media. Experiments 41-46 and 50-70 measure Rap1 occupancy at a single timepoint in the indicated strain during exponential growth in the presence of glucose. Enrichment in each deletion strain was compared to wildtype by t-test. B) For each strain the targets bound by Rap1 were determined by ChIPOTle35 (Methods). The percentage of low-glucose targets bound inappropriately by Rap1 is plotted. Error bars are not appropriate here because the percentage of targets bound is determined using all experimental replicates and takes measurement variability into account. Indicated at the bottom is the experiment number, the total number of new Rap1 targets observed relative to wild-type, the number of those new targets that were also low-glucose targets, and a statistic termed “specificity”, calculated as: (low-glucose targets / total new targets).