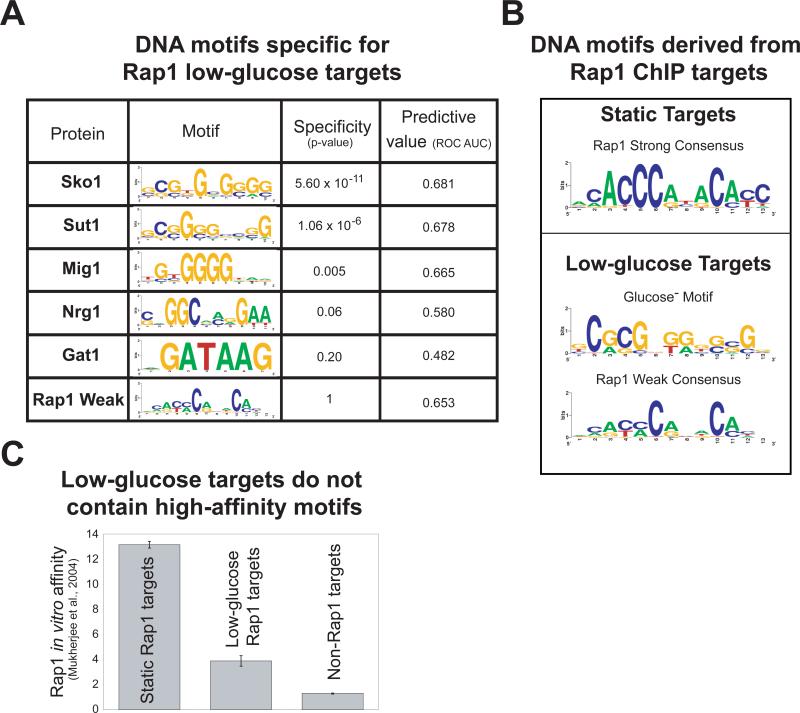
Figure 4. Low-glucose Rap1 targets contain Sko1, Sut1, or Mig1 binding sites and a weak Rap1 consensus motif.
A) The specificity of sequence motifs for low-glucose versus static targets was determined using ROVER41 with a site p-value cutoff of 0.001. The value of each motif in predicting Rap1 binding was determined using the area under (AUC) the receiver operator characteristic (ROC) plot42, with each promoter represented by the motif score generated by the MatrixScan module of BioProspector37. Motif scores encapsulate the similarity of DNA sequences at each locus to the specified position weight matrix (PWM). ROC plots show how low-glucose targets (true positives) were captured in relation to non-low-glucose targets (false positives) for a given motif score. A motif that had no predictive value would have an AUC of about 0.5; higher values are better (maximum = 1). B) Overrepresented DNA sequence motifs for static and low-glucose Rap1 targets were determined using MDscan and BioProspector37,38. The motif found for the static targets is the archetypical Rap1 binding motif. The most significant motif discovered for the low-glucose targets is very similar to the Sut1 binding motif2. We identified the Rap1-weak motif through a directed search using a degenerate Rap1-Strong PWM (methods). All PWMs are displayed as sequence logos40. C) Rap1 in vitro binding affinity for low-glucose-, static-, and non-Rap1 targets was enumerated using published protein binding microarray data29. “Protein binding microarray” is a technique that determines a protein's in vitro DNA-binding specificity for a set of arrayed loci or DNA sequences.