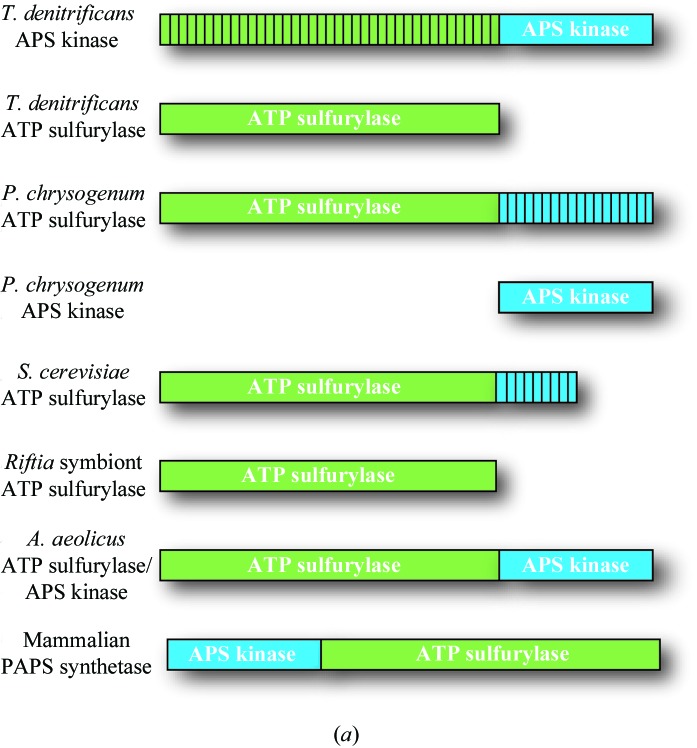
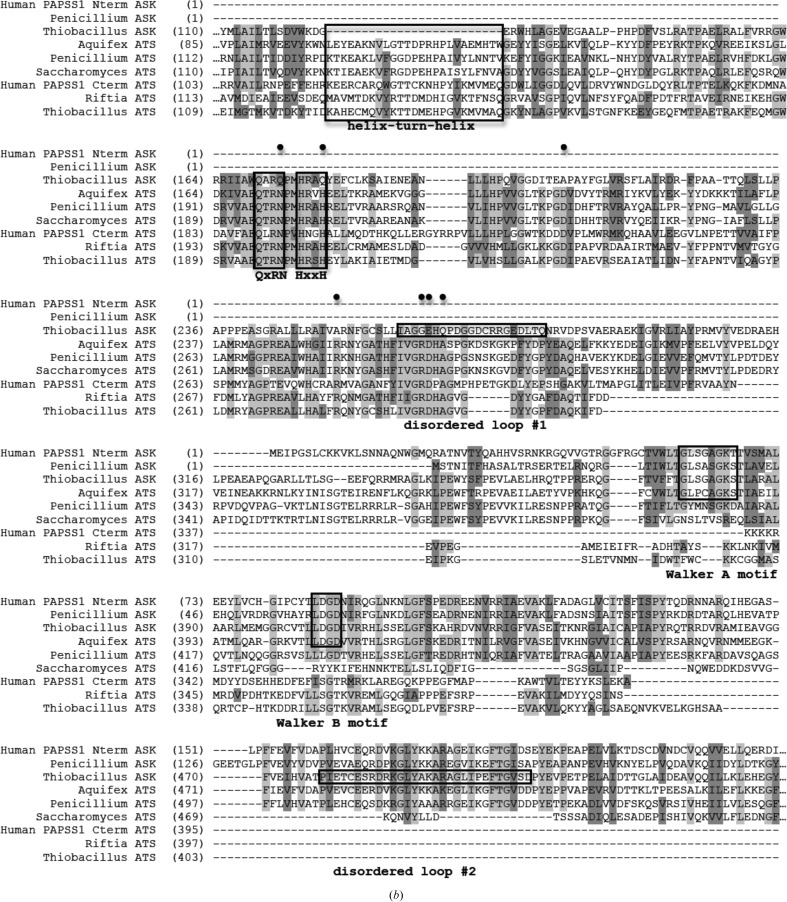
Figure 3.
Domain and sequence alignment of sulfur-activating enzymes from different organisms. (a) General domain arrangement of the ATP sulfurylase and APS kinase enzymes/domains from various species presented/discussed here. Green and blue represent ATP sulfurylase and APS kinase domains, respectively. Domains with vertical stripes indicate no enzymatic activity. (b) Sequence alignment. The bifunctional human PAPSS1 gene has been separated into two sequences owing to differences in enzyme architecture (its APS kinase domain precedes its ATP sulfurylase domain). Also shown are APS kinase and ATP sulfurylase from P. chrysogenum, APS kinase and ATP sulfurylase from T. denitrificans, the bifunctional ATP sulfurylase/APS kinase from A. aeolicus, ATP sulfurylase from S. cerevisiae and ATP sulfurylase from the Riftia symbiont. Black boxes denote the helix–turn–helix, QxRN, HxxH, Walker A and Walker B motifs as well as the two disordered loops in the Thiobacillus APS kinase structure. Black circles mark the locations of residues that are conserved in active ATP sulfurylase sequences but differ in the N-terminal domain of APS kinase from Thiobacillus.