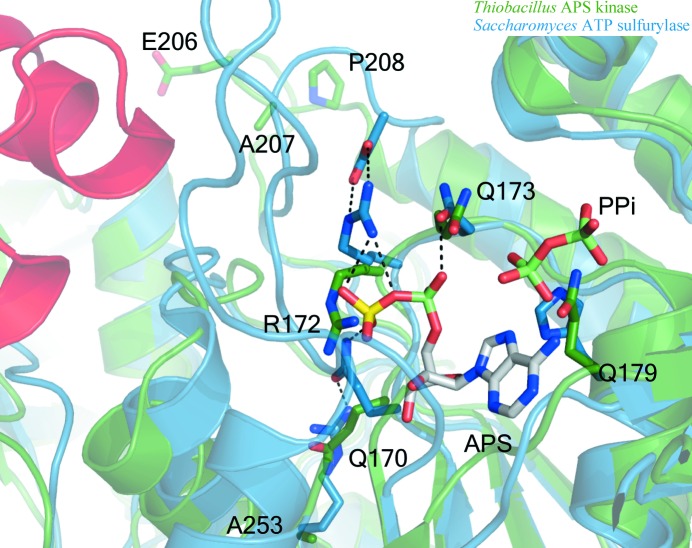
Figure 7.
Ribbon and stick diagram of ATP sulfurylase ‘active site’ overlay of T. denitrificans APS kinase (green) and S. cerevisiae ATP sulfurylase (blue, with the helix–turn–helix shown in red). T. denitrificans APS kinase shows poor orientation of essential ATP sulfurylase active-site residues when compared with ATP sulfurylase from S. cerevisiae. The QxRN-motif residues are in a particularly unfavorable position for ligand binding as a result of an Asn-to-Gln mutation at residue 173 and the mutation of secondary-layer residues (Arg to Ala253 and Asn to Ala207) that help to orient the active-site side chains. Pro208 is likely to stabilize the following helix, preventing it from unwinding to allow Glu206 from acting as a ‘surrogate’ Asp to orient Arg172. Additionally, a His-to-Gln mutation at residue 179 is likely to impede binding of the α- and β-phosphates of ATP. Ligands (APS and PPi) are modeled from the yeast structure (PDB code 1g8h). The helix–turn–helix motif that is missing from Thiobacillus is shown in red for perspective. Residue numbering is based on the Thiobacillus structure.