Figure 2.
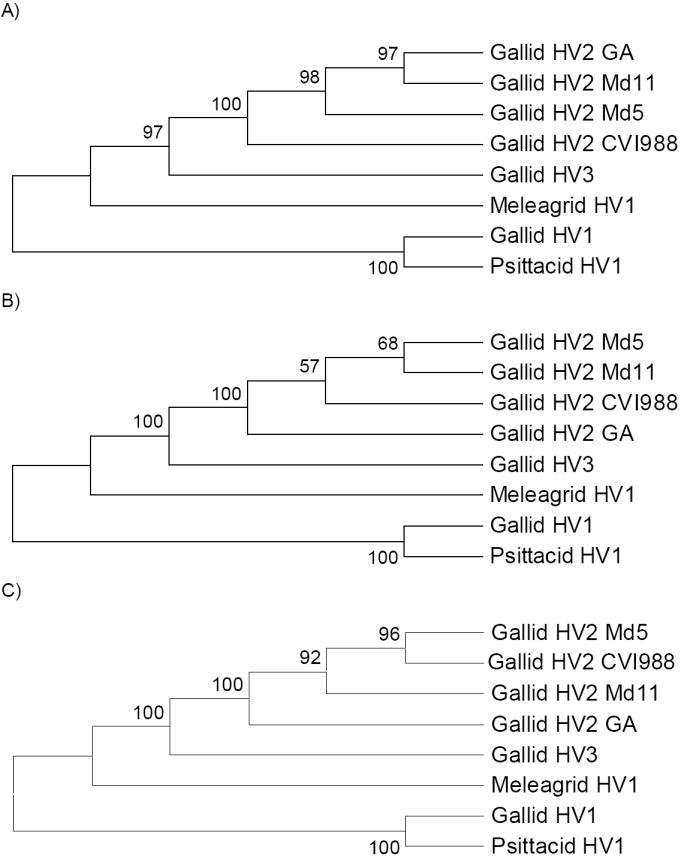
Phylogenetic trees (showing topology only) based on concatenated amino acid sequences of 31 orthologous genes from avian alphaherpesvirus genomes (15,476 aligned amino acid sites); trees are rooted by the mid-point method: (A) NJ tree based on the gamma distance (shape parameter = 1.16); (B) MP tree; and (C) QML tree based on the JTT with gamma (shape parameter = 1.16). Numbers on the branches are percentage of bootstrap samples supporting the branch (A and B) or percentage of puzzling steps supporting the branch (C).
