Figure 1.
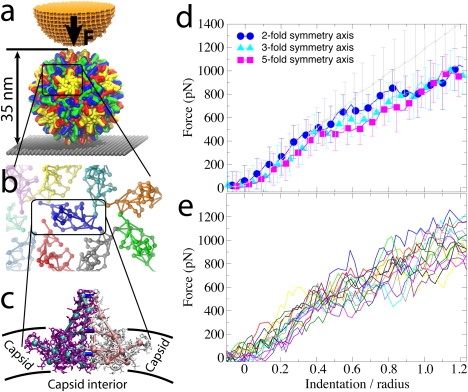
Experiments and CG simulations of AFM nanoindentations of the HBV T=4 capsid. (a) The capsid, composed of 240 identical protein units that form 120 homodimers, is attached to a surface (gray) and squeezed by the AFM tip (orange). (b) Arrangement of dimers (SBCG models) on the capsid surface, demonstrating their loose packing. (c) Within a dimer, the monomers (purple, white) are connected via extensive surface contacts. To model these contacts in the CG model, the monomers (cyan, pink) are connected by additional bonds (blue). (d) Force-indentation curves for the first round of AFM pushing, Zmax = 1.25R, averaged over all simulations for each pushing direction (color), and over all nanoindentation experiments (gray). Error bars are RMSD values. (e) Force-indentation curves for individual simulations with Zmax = 1.25R, each shown by a different color (data are averaged over 150-ns time windows).
