Figure 3.
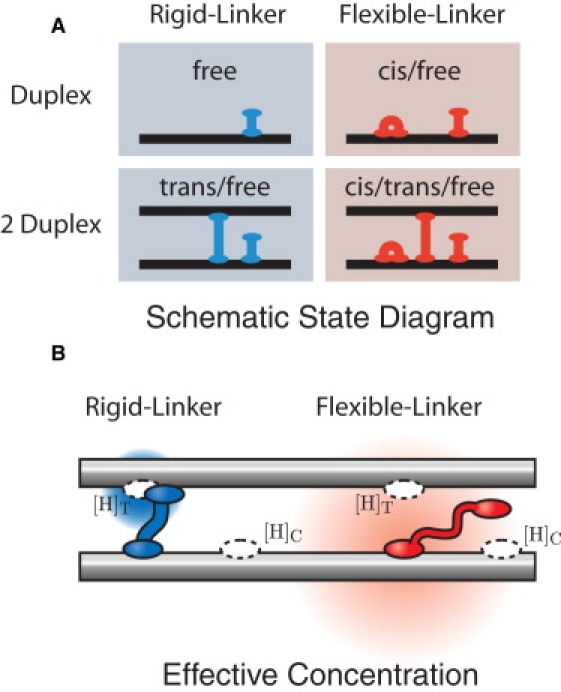
(A) Schematic state diagram of H-NS–DNA interactions. In our schematic drawings, we represent the H-NS dimer as a single unit. We consider two simple models for H-NS–DNA interaction: the Rigid and Flexible-Linker models. In the Flexible-Linker model, the H-NS dimer can bind to DNA in three states: free-head, trans, and cis. When H-NS interacts with two DNA duplexes, there is a competition between the three modes of binding. DNA adhesion is driven only by entropic effects (since H-NS can assume the cis conformation at the same energetic cost). In the Rigid-Linker model, the linker is too stiff to efficiently permit cis binding and the H-NS–DNA interaction is dominated by the free-head and trans binding modes. (B) The effective concentration illustrated for the Flexible and Rigid-Linker models. When H-NS binds a head to a DNA duplex, the diffusion of the second head is constrained. The physical concentration ([H]) of the second head in the proximal volume is dramatically increased. (The concentration is schematically illustrated by the red and blue gradients where the deeper hue corresponds to a higher local concentration.) This effective concentration is predicted by a polymer model for the linker domain. Structural and mutational studies of the H-NS protein suggest that the dimerization and DNA-binding domains are connected by a flexible linker (15). The Flexible-Linker model makes use of the fact that the linker domain can be modeled as a Gaussian chain with the generic amino-acid Kuhn length. In contrast, the linker domain of the Rigid-Linker model is assumed to have a structure which localizes the head domain in proximity to the trans-binding site. We therefore expect the trans head concentration to be much greater than that computed in the Flexible-Linker model.
