Figure 1.
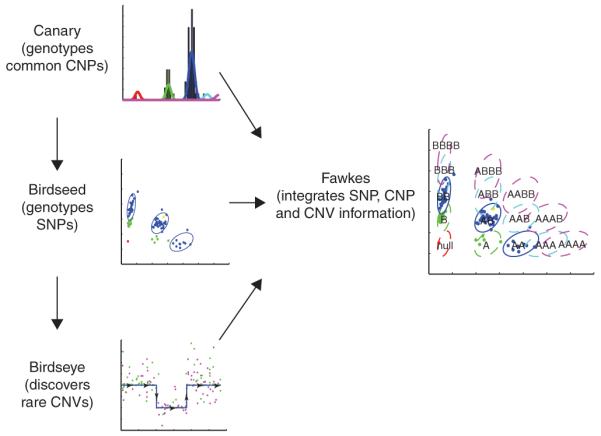
Overview of Birdsuite. In step 1, Canary estimates copy number across regions of known common copy number polymorphisms (‘CNP genotyping’). In step 2, Birdseed assigns canonical SNP genotypes (AA, AB or BB) to samples estimated by Canary to have two copies of a SNP locus (‘SNP genotyping’). Additionally, it calculates probe-specific mean and variances. In step 3, Birdseye estimates the likelihood of rare or de novo copy number variants, using probe-specific means and variances informed by Canary and Birdseed, and combining data across multiple probes in the region (‘CNV discovery’). In step 4, Fawkes combines copy number information for each sample at each locus with allele-specific information to assign a comprehensive SNP genotype, including noncanonical genotypes such as A-null or AAB.
