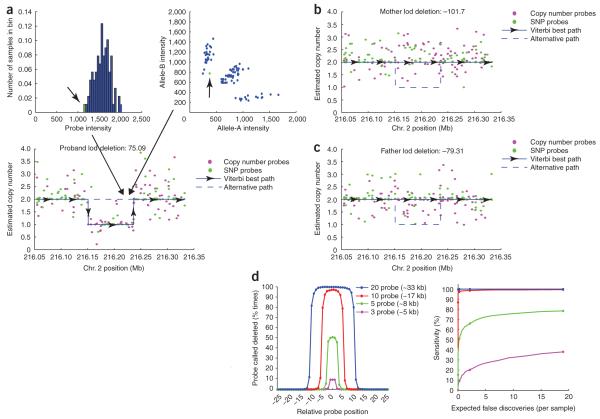
Figure 4.
Discovery of unknown or de novo copy number variation using Birdseye. (a) Raw data from a copy number probe, with one sample (arrow) colored green (top left). Raw data from a neighboring SNP, with the same sample (arrow) colored green (top right). Although the sample is relatively low in intensity, one would not have confidence calling a deletion on the basis of these data alone (bottom). A view across a larger region surrounding these two probe locations. A point is placed at the estimated copy number for this sample at each queried locus (without taking into account neighboring probes). With enough probes to support the evidence of a deletion, the HMM transitions to call a heterozygous deletion in this sample across an 85-kb region (blue line). (b,c) In addition, calling the deletion in the sample shown in a, Birdseye determines the relative log-likelihood of the identical deletion in each parent of this sample. Owing to strong evidence against this deletion in the parents, the region represents a de novo event in the child. (d) Data from in silico gender-mixing experiment. Sensitivity and breakpoint accuracy to discover simulated deletions of varying size (left). A deletion was considered discovered only if the lod score for the deletion was above 2. Sensitivity to discover the simulated deletions plotted against expected number of false-positive discoveries per genome (right). Points are placed at lod thresholds of 5, 2, 1 and 0.