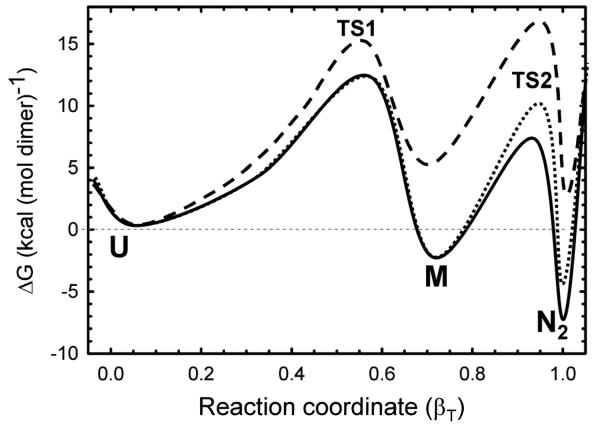
Figure 8.
Folding free energy surfaces for 10 μM (solid line) and 100 nM (dotted line) HIV-protease under native conditions (0 M urea) and for 10 μM (dashed line) HIV-PR* under unfolding conditions (4 M urea). The free energies were calculated for the 2U ⇆ 2M ⇆ N2 model using a dimeric reference state and the parameters listed in Table 1. The activation free energies for the transition states, ΔGo‡, were calculated from the rate constant, k, using the Kramers formalism, ΔGo=−RTln(k/ko), where R is the gas constant, T is the absolute temperature and ko is the Kramers prefactor. The activation energy depends upon the estimate of the prefactor and is somewhat arbitrary. The Kramers prefactor value of 1×108 s−1 used here is considered to be a reasonable estimate.54 The abscissa describes the reaction coordinate and is depicted as the Tanford β value, βT. The abscissa is normalized to the total change in the m value for the folding reaction from 2U to N2, 2.50 kcal (mol dimer)−1 M−1 (Table 1).