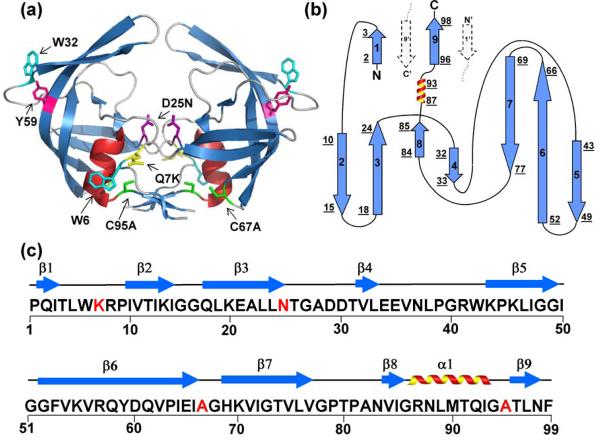
Figure 1.
Structure, topology and sequence of HIV-PR. (a) The crystal structure of unliganded, homodimer, D25N HIV-1 (1TW7) protease shows two β-barrel subunits with a dimerization interface comprised primarily of an anti-parallel β-sheet formed from the interdigitation of the N- and C- termini of each subunit. The flaps (residues 43-56) at the top of each barrel are in an “open position” with respect to each other. Mutated residues in the wild-type HIV-1 protease and FL chromophores are shown in stick model representation and are indicated in one subunit of the structure by residue number: Q7K (yellow), D25N (purple), C67A and C95A (green); intrinsic chromophores are indicated by Tyr59 (magenta) and Trp6 and Trp42 (cyan). (b) The topology map of one subunit has a jelly-roll β-barrel topology. β-strands are indicated with arrows, the α-helix is represented by a coil, and loop regions are displayed with lines. Strands are numbered successively, and the underlined numbers correspond to the N- and C-terminal residues of each secondary structural element. Dashed arrows indicate the N- and C- terminal β-strands from the interacting subunit. (c) The amino acid sequence of the HIV-1 protease variant used for these studies. Highlighted in red are the sites that were mutated to create the protease variant used in these studies, Q7K/D25N/C67A/C95A. The 9 β-strands and single α-helical secondary structural elements are indicated above the sequence.