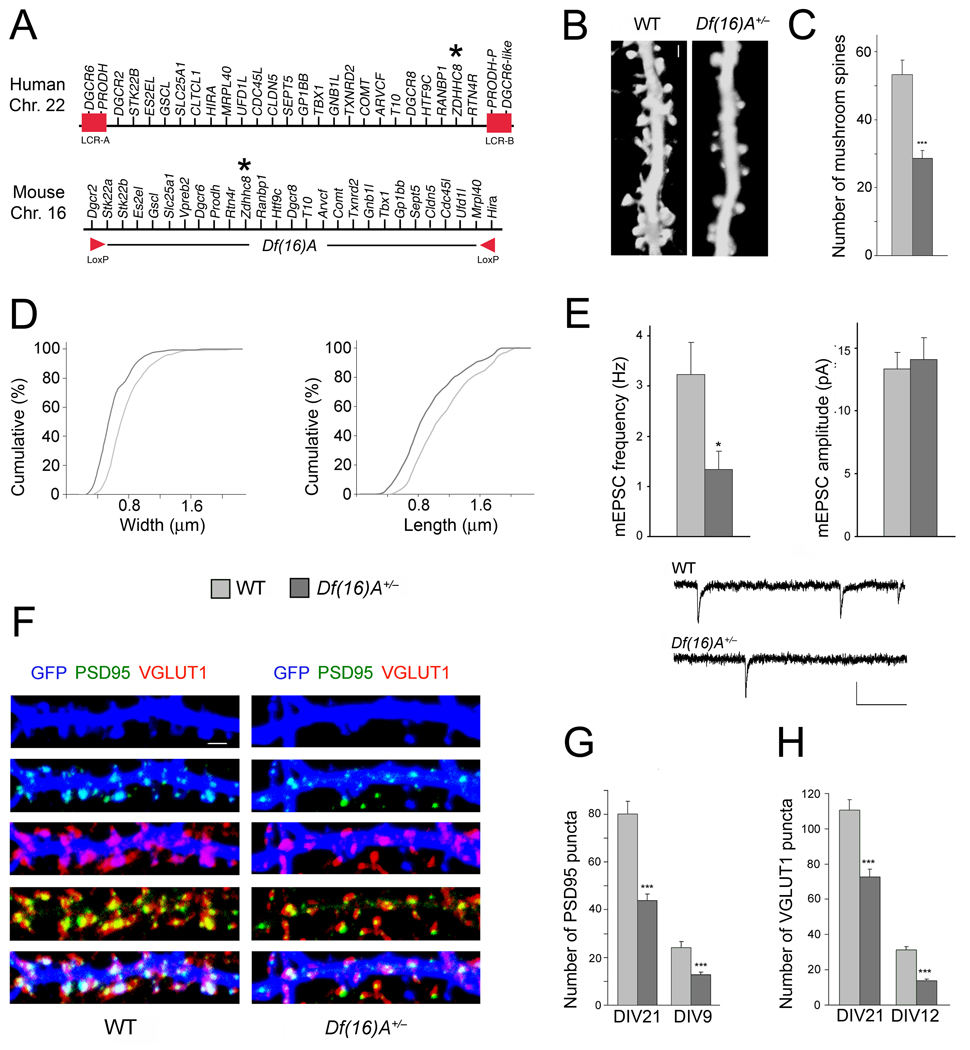
Figure 1. Df(16)A+/− neurons show reduced density of spines and glutamatergic synapses.
(a) Schematic showing the human 22q11.2 region and the syntenic mouse region. The 1.5-Mb deletion found in patients is mediated by low copy repeat sequences LCR-A and LCR-B (red boxes). Dgcr2 and Hira, the two endpoints of the targeted deletion where loxP sites were inserted23, are indicated by red arrowheads. The location of the human and mouse ZDHHC8 gene is indicated by an asterisk.
(b) Representative images of spines in Df(16)A+/− neurons (DIV21) transfected with a EGFP, which fills and clearly defines dendritic protrusions. Scale bar, 1 µm.
(c) Reductions in the number of mushroom spines observed at DIV21 on Df(16)A+/− neurons (estimated over 75 µm of dendritic length).
(d) Reductions in the width (left) and length (right) of mushroom spines on Df(16)A+/− neurons. P < 0.05, K-S test.
(e) Properties of mEPSC recorded from autaptic individual pyramidal neurons grown on astrocytic microislands. Top left: the frequency at which events occurred was lower in Df(16)A+/− neurons (n = 10) than in WT controls (n = 11). Top right: The average amplitude of mEPSCs was not different between genotypes. Bottom: Traces of recordings from a WT and a Df(16)A+/− neuron. Scale bars, vertical: 20 pA, horizontal: 100 msec.
(f) Representative images of PSD95 puncta (green) and VGLUT1 puncta (red) and their co-localization in GFP (blue)-labeled hippocampal neurons at DIV21. Scale bar, 1 µm.
(g) Reduction in the number of PSD95 puncta in Df(16)A+/− neurons at DIV21 and DIV9.
(h) Reduction in the number of VGLUT1 puncta in Df(16)A+/− neurons at DIV21 and DIV12. Data are shown as mean ± S.E.M. * P < 0.05, *** P < 0.0001.