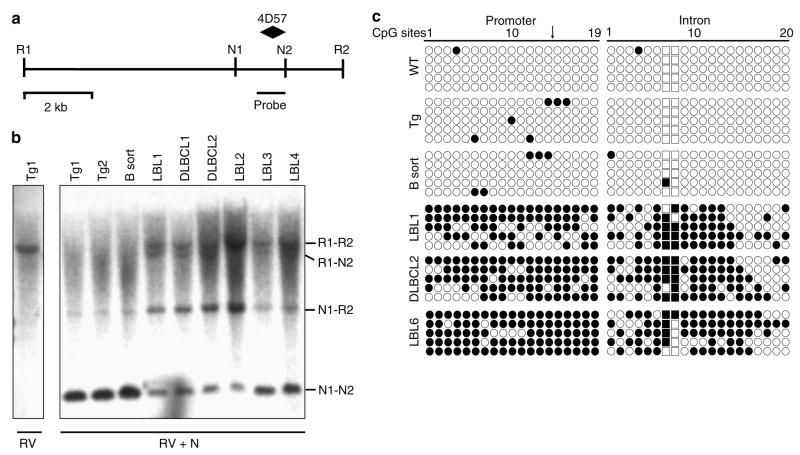
Figure 2.
Validation of promoter methylation for Epha7. (a) Schematic diagram of genomic EcoRV–EcoRV fragment (sites R1 and R2) of murine Epha7 containing two NotI sites (N1 and N2), as well as locations of the 4D57 NotI-HinfI RLGS fragment and Southern probe. (b) Southern hybridization for Epha7 on genomic DNA from non-malignant TCL1-tg spleens (Tg) or sorted B-cells (B sort) and malignant B-cell tumors. Digestions with EcoRV alone (first lane, RV) or in combination with methyl-sensitive NotI (remaining lanes, RV+N1) are indicated. Detected bands correspond to methylation of neither (N1–N2), both (R1–R2) or one of two NotI sites (R1–N2 or N2–R2). (c) Bisulfite sequence analysis of CpG islands located in the 5′ UTR and first intron of Epha7. Each row represents the sequence of an individual clone (open circle, unmethylated CpG site; filled circle, methylated site; squares, CpGs in the NotI site of the 4D57 locus; arrow, location of translational start site).