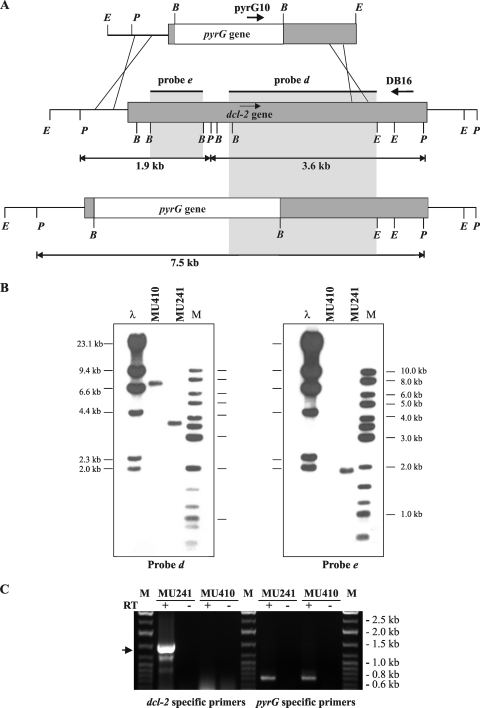
FIG. 5.
Disruption of the dcl-2 gene. (A) Schematic representation of the wild-type dcl-2 locus (middle) and after homologous recombination with the disruption fragment (below). Dark-gray boxes, genomic dcl-2 loci; white boxes, pyrG selectable markers. The positions of the probes used and the expected sizes of the PstI restriction fragments are indicated. P, PstI; E, EcoRI; B, BamHI. The primers used to identify homologous integration events are shown (pyrG10 and DB16). (B) Southern blot analysis of the dcl-2+ strain MU241 and the dcl-2− mutant MU410. Genomic DNA (1 μg) was digested with PstI and hybridized with probe d, which corresponds to a 2.5-kb XhoI-EcoRI fragment of the dcl-2 coding region (left), and with probe e, a 1.2-kb BamHI fragment isolated from plasmid pMAT1208 (right). The positions and sizes of the λ-HindIII (λ) and the GeneRuler DNA ladder mixture (M) (Fermentas) size markers are indicated. (C) RT-PCR analysis of dcl-2 expression in the dcl-2− mutant. Total RNA isolated from the wild-type strain MU241 and the dcl-2− mutant MU410 was retrotranscribed using an oligo(dT) primer and amplified with primers corresponding to dcl-2 sequences flanking the RNase III catalytic domains (left side). The arrow marks the size of the expected fragment. As a positive control, pyrG-specific primers were used to amplify pyrG transcripts (right side). Total RNA without RT was used as a negative control. M, GeneRuler DNA ladder mixture (Fermentas).