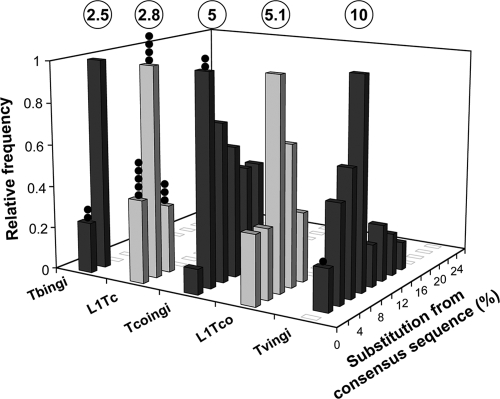
FIG. 1.
Divergence between members of potentially active retroposons belonging to the ingi clade. Bases covered by the T. brucei (Tbingi), T. cruzi (L1Tc), T. congolense (Tcoingi and L1Tco), and T. vivax (Tvingi) retroposons were sorted by their divergence from their consensus sequences. The consensus sequences were determined from the alignment of full-length retroposons (Tbingi, 63 copies; L1Tc, 48 copies; Tcoingi, 27 copies) and all of the annotated elements larger than 300 bp (L1Tco, 8 copies) or 3.5 kb (Tvingi, 46 copies). The number of retroposons per fraction of 2% divergence is expressed as a fraction of the highest value, to which an arbitrary value of 1 was assigned. The percentage of divergence was calculated using the matching regions of the consensus sequences. The circled numbers at the top indicate the median value of each graph, and the closed circles represent the number of potentially active elements in each column.