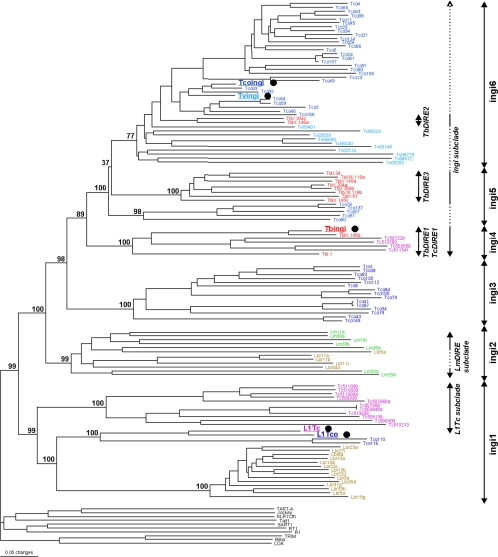
FIG. 3.
Phylogenetic analysis of the RT domain. The phylogeny is based on approximately 450 aligned amino acid residues (see Fig. S1 in the supplemental material for the alignment) corresponding to the entire RT domain of nontrypanosomatid retroposons, Tbingi, Tcoingi, Tvingi, L1Tc, L1Tco, and DIREs, which all belong to the ingi clade. The names of retroposons from T. brucei, T. congolense, T. vivax, T. cruzi, L. major, and L. braziliensis are in red, dark-blue, light-blue, magenta, green, and brown letters, respectively. The 10 nontrypanosomatid retroposons are representatives of the Jockey, Tad1, R1, CR1, and LOA retroposon clades (13). The consensus tree was generated by the neighbor-joining method and rooted on the RT sequences of nontrypanosomatid retroposons. The number next to each node indicates the bootstrap value as a percentage of 100 replicates corresponding to the tree generated by the neighbor-joining method. The black dots indicate potential functional autonomous retroposons. The names in italics on the right correspond to groups or subclades identified in a previous analysis (7), while the ingi1 to -6 subclades were defined from this phylogenetic analysis.