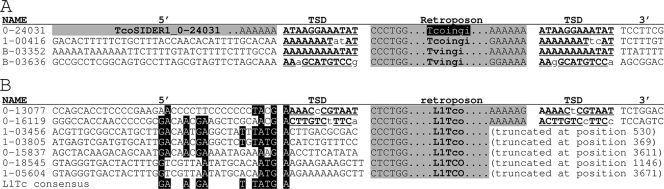
FIG. 6.
Comparison of the 5′ and 3′ adjacent sequences flanking the Tvingi and Tcoingi retroposons. (A) Only full-length Tcoingi and Tvingi flanked by a TSD presenting two mismatches at the most are considered. (B) The 5′ adjacent regions of L1Tco elements, which contain residues of the conserved motif located at the same relative position upstream of the T. cruzi L1Tc retroposon (4). Conserved residues are indicated by white letters on a black background. On the right, the names of the elements are indicated. The Tcoingi or L1Tco numbers with six characters correspond to the scaffold number (0 or 1) followed by the position (without the last three numbers) of the retroposon in the T. congolense scaffold. For the T. vivax retroposons (Tvingi), the numbers correspond to the contig numbers. The alignment of all the selected sequences was based on the retroposon sequences (gray column with “Tcoingi,” “Tvingi,” or “L1Tco”), from which only the first and last 6 bp, separated by the type of retroposon, are shown. The potentially functional Tcoingi elements that code for a full-length protein (1,751 amino acids) are indicated by white characters on a black background. The TSD flanking the retroposons is indicated by boldface letters, with underlined capital letters for the conserved residues; the lowercase letters in the TSD correspond to nonconserved residues. A gray background in the 5′ adjacent sequences (5′) means that the retroposon is preceded by another retroposon. For truncated retroposons, the position of end of the element is given.