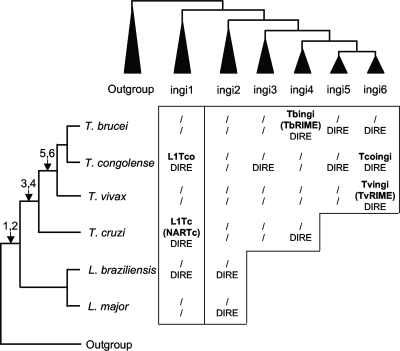
FIG. 9.
Evolution of ingi retroposons in the trypanosomatid genomes. On the left is shown the phylogenetic relationship between trypanosomatid species and subspecies analyzed here. The tree was drawn according to data from Stevens et al. (36) and Gibson et al. (16). The arrows indicate the deduced times of appearance of the ingi1 to -6 subclades. At the top is a schematic representation of the retroposon phylogenetic tree shown in Fig. 3, and the central table shows the retroposons belonging to the ingi1 to -6 subclades identified in the genomes of these trypanosomatids. The names of potential active ingi-related retroposons are in boldface. Also indicated are vestiges of ingi-like retroposons (DIRE) and, in parentheses, the short nonautonomous retroposons (TbRIME, TvRIME, and NARTc) corresponding to active truncated versions of the long and autonomous elements mentioned above (Tbingi, Tvingi, and L1Tc, respectively). The slashes indicate loss of the active retroposon and/or the corresponding vestigial sequences.