FIG. 6.
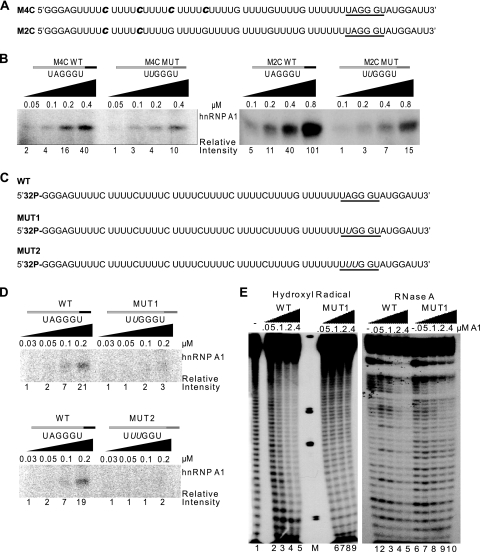
Mapping the extent of hnRNP A1 cooperative spreading along the RNA. (A) Sequences of synthetic RNA transcripts for UV cross-linking experiments. The radiolabeled cytidines are indicated in bold italic. (B) UV cross-linking of the 16 to 32 nM RNA transcripts from panel A and the corresponding ESSm controls, in the presence of increasing recombinant hnRNP A1 (0.05 to 0.8 μM). Left panel, cross-linked products after RNase digestion of the top transcript (16 nM) in panel A and its ESSm counterpart. Right panel, Idem for the bottom transcript (32 nM) in panel A and its ESSm counterpart. Band intensities were measured on a phosphorimager, and normalized values are shown below the gel. (C) Sequences of 5′-end-labeled synthetic RNA transcripts for UV cross-linking and footprinting experiments. The underlined hexanucleotide is a high-affinity hnRNP A1 binding site: UAGGGU is the WT version, UUGGGU is the inactive mutant version 1 (MUT1), and UUUGGU is the inactive mutant version 2 (MUT2), with the mutated nucleotide shown in italic. (D) UV cross-linking with 8 nM WT and MUT1 and MUT2 RNAs from panel C in the presence of increasing concentrations of recombinant hnRNP A1 (0.03 to 0.2 μM). Detection and quantitation of the cross-linked products were as for Fig. 1B. (E) Footprinting assays with the first two RNA transcripts from panel C (WT and MUT1). The left panel shows a hydroxyl radical footprinting assay, and the right panel shows an RNase A footprinting assay; both were carried out with 16 nM 5′-end-labeled RNA in the presence of increasing recombinant hnRNP A1 (0 to 0.4 μM). M, molecular weight markers.