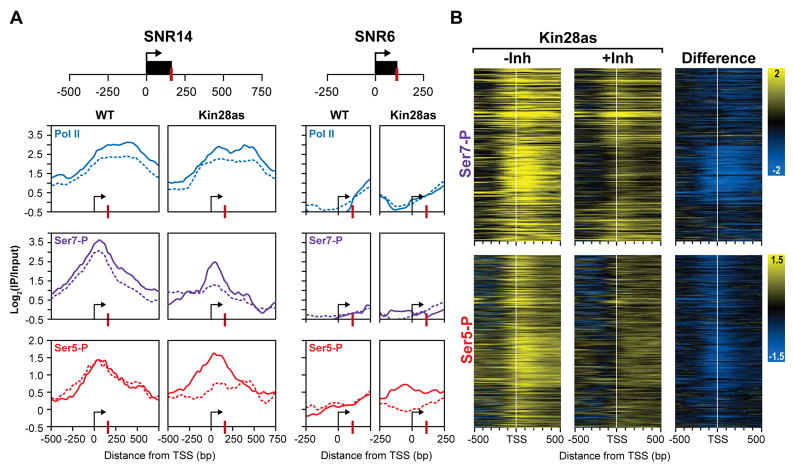
Figure 3. In vivo inhibition of Kin28 affects the Ser7 phosphorylation pattern.
(A) ChIP-chip profiles for two snRNAs (remaining three are in the supplemental figures). The occupancy profiles of Pol II (blue), Ser7-P (purple) and Ser5-P (red) at SNR14 and SNR6 (control) are shown. Uninhibited profiles are shown as solid lines and profiles in chemically inhibited cells are shown as dashed lines. TSS and 3′ processing sites are marked by an arrow and a red bar respectively. All x-axes are shown as the distance in base pairs relative to the TSS and y-axes are shown on a log2 scale for ChIP-chip enrichment (IP/input).
(B) Heat maps of 308 protein-coding genes showing Ser7-P and Ser5-P occupancy profiles over a 1kb region centered over the TSS (white bar) for the Kin28as strain with and without inhibition. The change in profiles upon inhibition is shown on the right. Yellow profiles indicate enrichment and blue profiles indicate depletion of the phosphorylated CTD.