Abstract
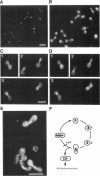
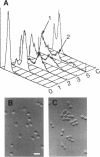
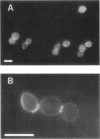
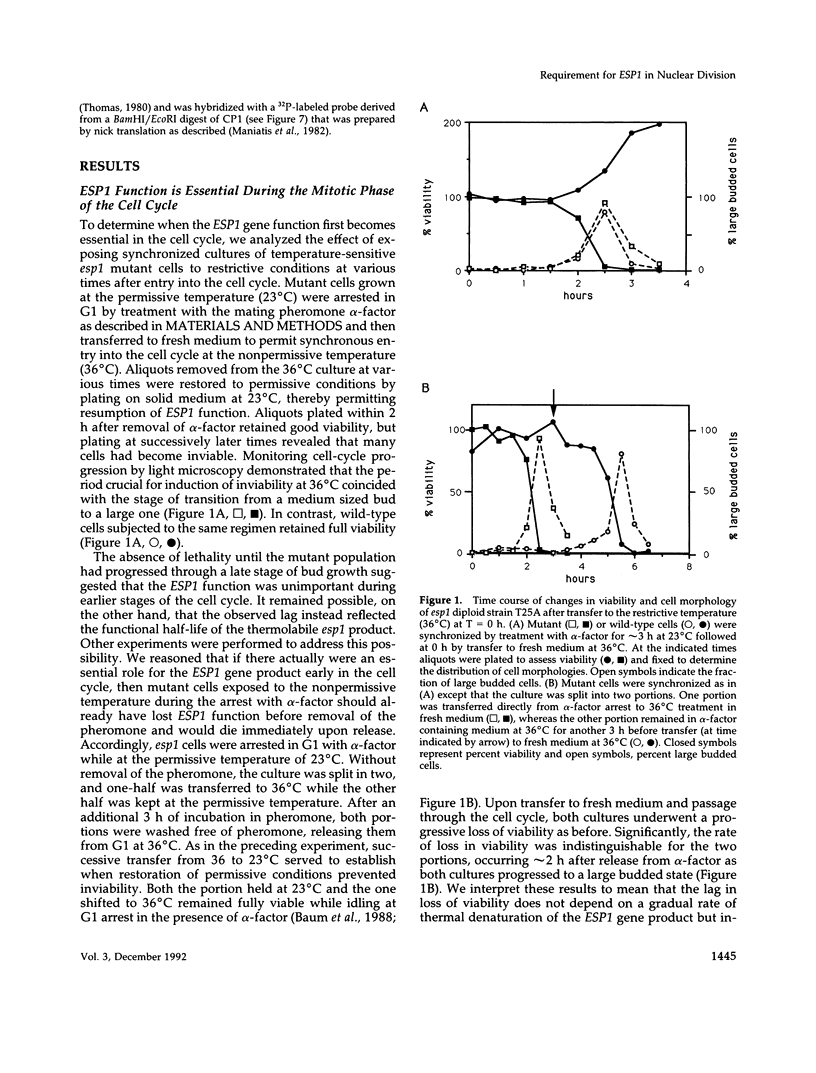
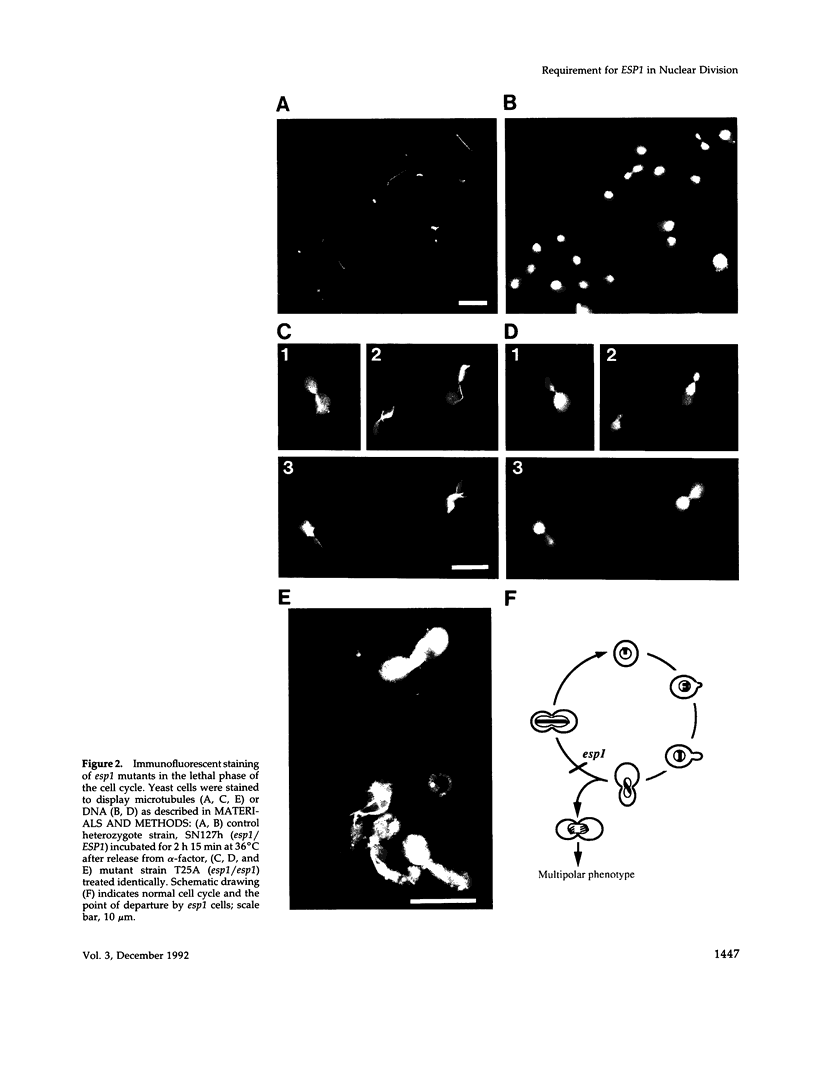
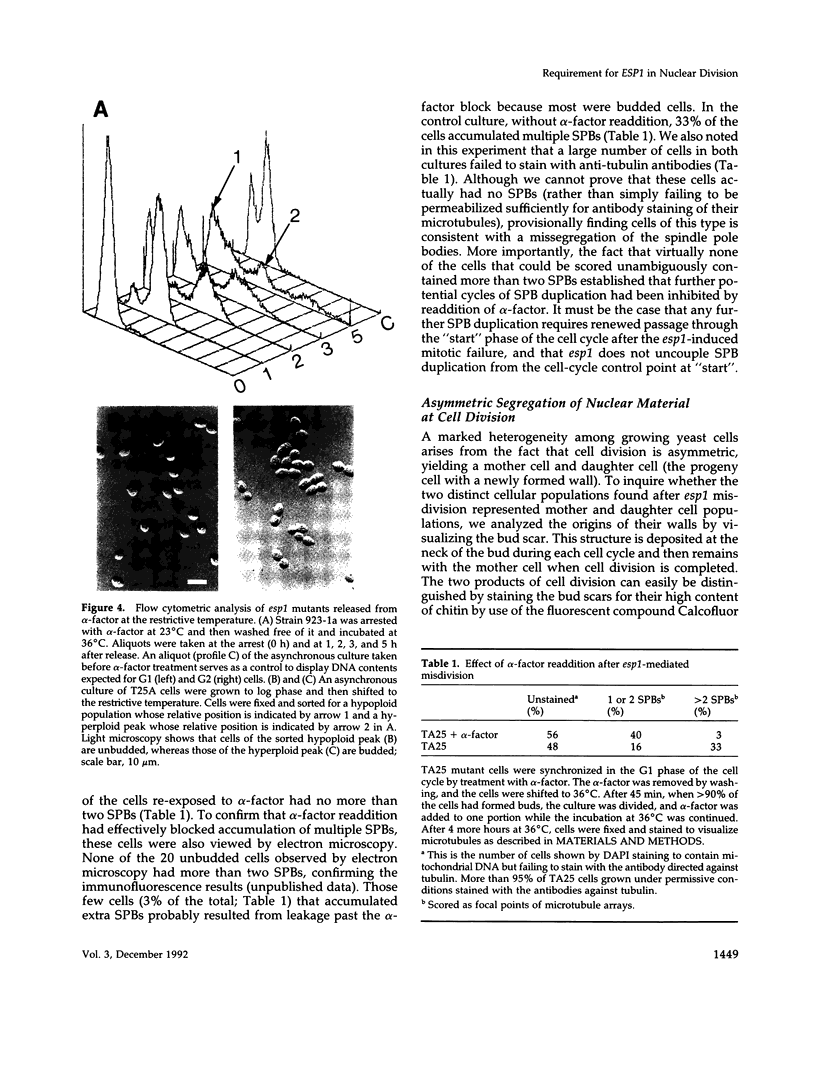
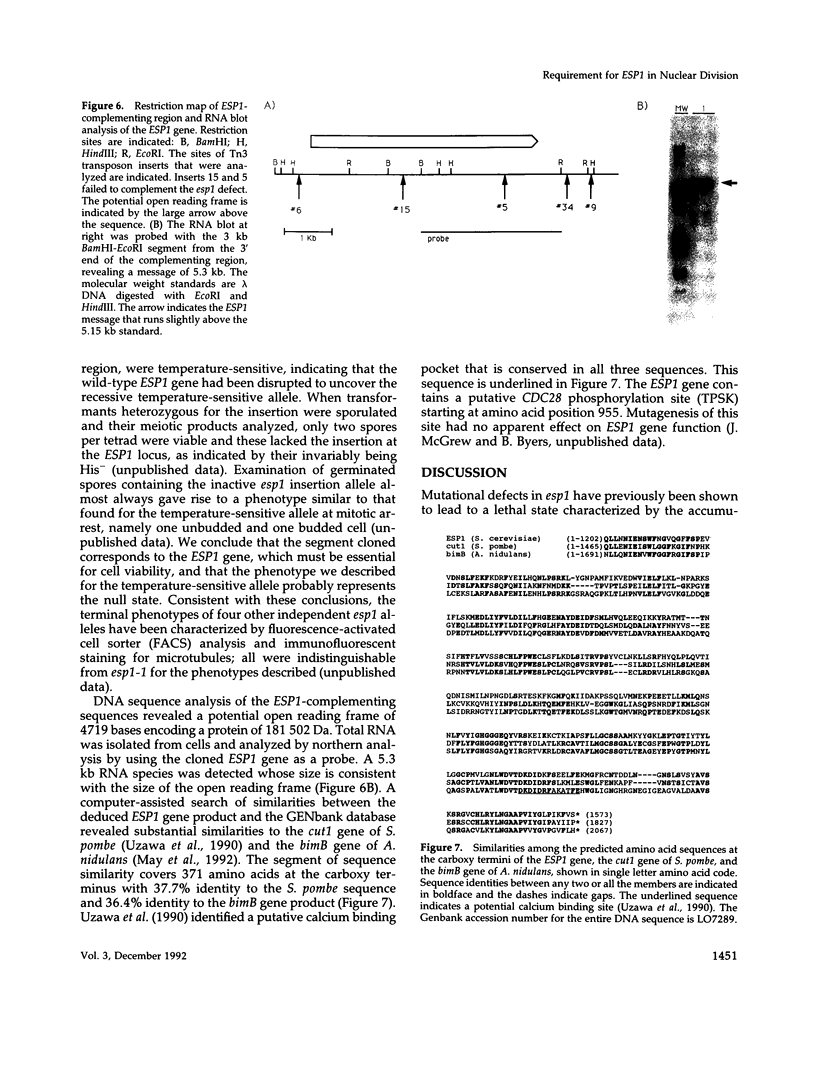
Mutations in the ESP1 gene of Saccharomyces cerevisiae disrupt normal cell-cycle control and cause many cells in a mutant population to accumulate extra spindle pole bodies. To determine the stage at which the esp1 gene product becomes essential for normal cell-cycle progression, synchronous cultures of ESP1 mutant cells were exposed to the nonpermissive temperature for various periods of time. The mutant cells retained viability until the onset of mitosis, when their viability dropped markedly. Examination of these cells by fluorescence and electron microscopy showed the first detectable defect to be a structural failure in the spindle. Additionally, flow cytometric analysis of DNA content demonstrated that massive chromosome missegregation accompanied this failure of spindle function. Cytokinesis occurred despite the aberrant nuclear division, which often resulted in segregation of both spindle poles to the same cell. At later times, the missegregated spindle pole bodies entered a new cycle of duplication, thereby leading to the accumulation of extra spindle pole bodies within a single nucleus. The DNA sequence predicts a protein product similar to those of two other genes that are also required for nuclear division: the cut1 gene of Schizosaccharomyces pombe and the bimB gene of Aspergillus nidulans.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Yip C., Goetsch L., Byers B. A yeast gene essential for regulation of spindle pole duplication. Mol Cell Biol. 1988 Dec;8(12):5386–5397. doi: 10.1128/mcb.8.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Creanor J., Mitchison J. M. Continued DNA synthesis after a mitotic block in the double mutant cut1 cdc11 of the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1990 Jul;96(Pt 3):435–438. doi: 10.1242/jcs.96.3.435. [DOI] [PubMed] [Google Scholar]
- Cross F., Hartwell L. H., Jackson C., Konopka J. B. Conjugation in Saccharomyces cerevisiae. Annu Rev Cell Biol. 1988;4:429–457. doi: 10.1146/annurev.cb.04.110188.002241. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Hepler P. K. Calcium transients during mitosis: observations in flux. J Cell Biol. 1989 Dec;109(6 Pt 1):2567–2573. doi: 10.1083/jcb.109.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Funahashi S., Uemura T., Yanagida M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986 Nov;5(11):2973–2979. doi: 10.1002/j.1460-2075.1986.tb04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hogan C. J., Cande W. Z. Antiparallel microtubule interactions: spindle formation and anaphase B. Cell Motil Cytoskeleton. 1990;16(2):99–103. doi: 10.1002/cm.970160203. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985 Jun;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991 Aug 9;66(3):507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Huffaker T. C., Thomas J. H., Botstein D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J Cell Biol. 1988 Jun;106(6):1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K. J., Eipel H. E. Microbial determinations by flow cytometry. J Gen Microbiol. 1979 Aug;113(2):369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C. W., Adams A. E., Szaniszlo P. J., Pringle J. R. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988 Oct;107(4):1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991 Aug 9;66(3):519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Maccecchini M. L., Rudin Y., Blobel G., Schatz G. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc Natl Acad Sci U S A. 1979 Jan;76(1):343–347. doi: 10.1073/pnas.76.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May G. S., McGoldrick C. A., Holt C. L., Denison S. H. The bimB3 mutation of Aspergillus nidulans uncouples DNA replication from the completion of mitosis. J Biol Chem. 1992 Aug 5;267(22):15737–15743. [PubMed] [Google Scholar]
- Moore S. A. Synchronous cell growth occurs upon synchronizing the two regulatory steps of the Saccharomyces cerevisiae cell cycle. Exp Cell Res. 1984 Apr;151(2):542–556. doi: 10.1016/0014-4827(84)90402-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Palmer R. E., Koval M., Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989 Dec;109(6 Pt 2):3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Adams A. E., Drubin D. G., Haarer B. K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. H., Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986 Jan 17;44(1):65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa S., Samejima I., Hirano T., Tanaka K., Yanagida M. The fission yeast cut1+ gene regulates spindle pole body duplication and has homology to the budding yeast ESP1 gene. Cell. 1990 Sep 7;62(5):913–925. doi: 10.1016/0092-8674(90)90266-h. [DOI] [PubMed] [Google Scholar]
- Weinert T. A., Hartwell L. H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988 Jul 15;241(4863):317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991 Aug;114(4):745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]