Abstract
Herpes simplex virus type 1 encodes a multifunctional protein, ICP8, which serves both as a single strand binding protein and recombinase, catalyzing reactions involved in replication and recombination of the viral genome. In the presence of divalent ions and at low temperature, previous electron microscopic (EM) studies showed that ICP8 will form long left-handed helical filaments. Here EM image reconstruction reveals that the filaments are bipolar, with an asymmetric unit containing two subunits of ICP8 that constitute a symmetrical dimer. This organization of the filament has been confirmed using Scanning Transmission Electron Microscopy. The pitch of the filaments is ~ 250 Å, with ~ 6.2 dimers per turn. Docking of a crystal structure of ICP8 into the reconstructed filament shows that the C-terminal domain of ICP8, attached to the body of the subunit by a flexible linker containing ~ 10 residues, is packed into a pocket in the body of a neighboring subunit in the crystal in a similar manner as in the filament. However, the interactions between the large N-terminal domains are quite different in the filament from that observed in the crystal. A previously proposed model for ICP8 binding single-stranded DNA, based upon the crystal structure, leads to a model for a continuous strand of ssDNA near the filament axis. The bipolar nature of the ICP8 filaments means that a second strand of ssDNA would be running through this filament in the opposite orientation, and this provides a potential mechanism for how ICP8 anneals complementary single stranded DNA into double-stranded DNA, where each strand runs in opposite directions.
Keywords: Herpes simplex virus, ICP8, electron microscopy, image analysis, filament
Introduction
Herpes Simplex Virus type 1 provides a powerful system for studies of DNA metabolism in eukaryotic cells. Seven viral encoded proteins required for reconstitution of replication in vitro have been identified, purified, and characterized in detail1. One of these, ICP8, is of particular interest due to its unique multifunctional properties and the presence of direct counterparts in other Herpes viruses including CMV, EBV and KSHV2.
ICP8 is a 128 kDa globular zinc metalloprotein expressed in abundance in HSV-1 infected cells3,4. It was originally identified as a single strand DNA (ssDNA) binding protein and assumed to function like E. coli SSB or eukaryotic RPA in binding ssDNA generated during replication4-6. ICP8 binds ssDNA cooperatively7,8 and holds it in an extended conformation9,10. ICP8 facilitates replication in vitro by the HSV-1 DNA polymerase complex UL30/4211,12. ICP8, however, was found to have additional activities not associated with the classical SSBs. It forms a specific co-complex with the origin binding protein UL9 that functions in opening and activating the HSV-1 oriS origin13-16. ICP8 also functions as a single strand annealing protein in catalyzing the annealing of complementary ssDNA17,18. It also shares properties with some of the recombinases as it will promote an invasion of ssDNA into dsDNA and can catalyze strand transfer between ssDNA and homologous dsDNA molecules containing ssDNA tails19-21. If homologous dsDNA molecules with resected ends are provided, ICP8 is able to drive strand transfer over 7 kb in an ATP-independent reaction22,23. ICP8 is involved in the nuclear compartmentalization of replicating HSV-1 DNA24, stimulates late gene transcription, and is associated with the RNA polymerase II holoenzyme in infected cells25.
The formation of helical ICP8-DNA filaments is central to some, if not all, of these reactions. However, the similarity of these nucleoprotein filaments to those formed by bacterial RecA, and archaeal and eukaryotic RecA-like proteins, such as RadA26 and Rad5127, has not been clear. Like RecA, ICP8 protein alone will assemble into helical protein filaments that resemble ones formed on DNA. These self filaments are formed when ICP8 is incubated in the presence of Mg2+ at low temperature (4°C)12. When ICP8 binds to ssDNA it holds the DNA in an extended conformation with a 3.6 Å axial rise between adjacent nucleotides9,10, similar to the 3.4 Å rise in dsDNA. The conformation of these ssDNA-ICP8 filaments is highly dependent on the presence of divalent cations, and in the presence of Mg2+ the helical filaments intertwine to form superhelices18. Such superhelices were shown to promote the annealing of complementary ssDNA18,28.
To understand the mechanisms by which ICP8 functions in its many roles, it will be essential to understand the structure of the filaments which it forms both alone and in the presence of DNA. A great deal of insight into how proteins such as RecA function has come from structural studies using both electron microscopy29 and x-ray crystallography30. In the present study we have used three-dimensional reconstruction of the filament which ICP8 forms in the absence of DNA. We show that the filaments have about 6.2 asymmetric units per helical turn, but the asymmetric unit is a dimer of ICP8. The filaments are bipolar since the dimer axis is perpendicular to the filament axis. The predicted mass per unit length of these filaments is in good agreement with direct mass analysis carried out using Scanning Transmission Electron Microscopy (STEM). A previously proposed model for how each subunit of ICP8 binds ssDNA leads to an interesting model for how ICP8 functions in the annealing of two strands of DNA, each running in an opposite direction.
TEM visualization of ICP8 filaments and analysis of heterogeneity
Previous observations have shown that HSV-1 ICP8 forms a left-handed helical filament in the presence of Mg2+ at low temperature12,31. ICP8 (HSV-1 strain 17) was purified from Sf9 insect cells infected with Baculovirus construct (a gift from Dr. Nigel D. Stow, Institute of Virology, Glasgow, UK) as described32. The filaments were formed by incubation of ICP8 in buffer containing 20 mM Tris-HCl, pH 7.5, 50 mM NaCl in the presence of 5 mM MgCl2 at 4°C for 12 – 16 h. The negatively-stained ICP8 filaments (Fig. 1a) showed structural heterogeneity, as evidenced by a variable helical pitch (data not shown). To determine if this variability might arise as a consequence of specimen preparation, we imaged unstained frozen-hydrated filaments in ice using cryo-EM (Fig. 1b). A sorting of 4011 segments based upon pitch, using a cross-correlation against projections of reference volumes having different pitches, shows (Fig. 1c) the very large variability in pitch observed in ice. Since the variability in ice was comparable to that seen by negative stain, we have used the negatively-stained filaments, with their much higher signal-to-noise ratio, for subsequent analysis and processing.
Fig. 1.
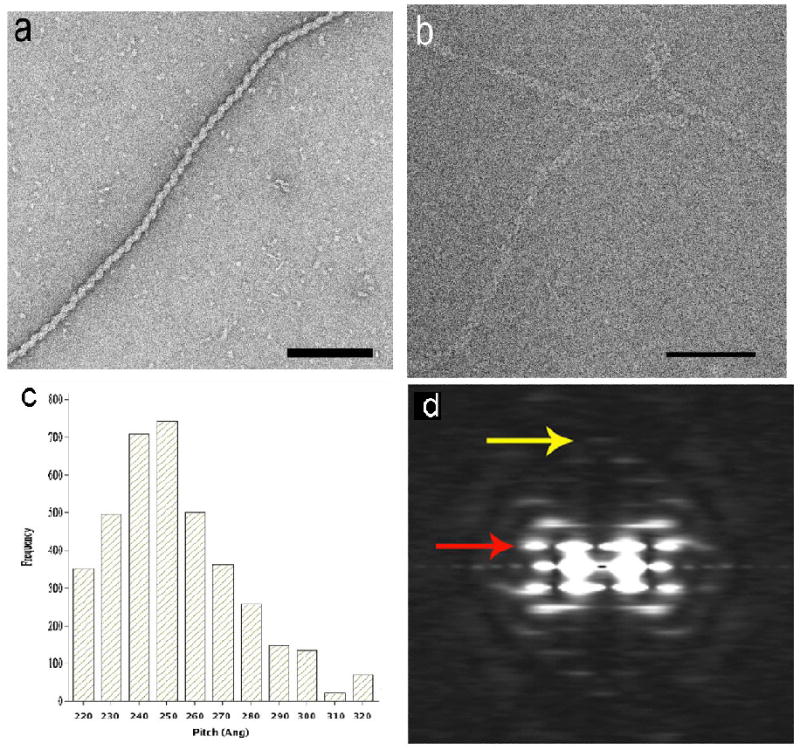
Electron micrograph of a negatively-stained ICP8 filament (a), and unstained frozen-hydrated ICP8 filament (b). ICP8 protein (50 μg/ml) in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2 incubated for 8 – 16 h at 4° C were adsorbed directly onto glow-charged thin carbon foils without fixation and stained with 2% (w/v) uranyl acetate. The grids were imaged in a Philips CM12 TEM at an accelerating voltage of 80kV under minimal-dose conditions and recorded on Kodak SO-163 films at a magnification of 45K. Cryo-EM images were recorded in Tecnai 20 with field emission gun at a magnification of 50K at an accelerating voltage of 200 kV after incubation of 1 μM ICP8 in the above indicated buffer for 2 – 3 weeks in ice. The variability in the pitch of the frozen-hydrated ICP8 filaments (c) was determined by sorting 4011 filament segments using reference models having pitches from 220 to 320 Å. An average power spectrum, generated from 181 segments (ranging in length from 150 to 400 pixels) of the negatively-stained filaments (d) shows a meridional layer line (yellow arrow) at ~ 1/(40 Å) that is the reciprocal of the axial rise per subunit. The strongest layer line (red arrow), at ~ 1/(250 Å), arises from the one-start helix.
An averaged power spectrum generated from 181 long individual negatively-stained ICP8 filaments (ranging in length from 900 Å to 1,200 Å) showed a clear meridional reflection (yellow arrow, Fig. 1d) at ~ 1/(40 Å) indicating that the axial rise per subunit within these filaments was ~ 40 Å. A layer line (red arrow, Fig. 1d) is seen at ~ 1/(250 Å) that can only arise from a one-start helix, given the distance of the peaks from the meridian and the diameter of the filaments (~ 180 Å). These two observations completely define the helical symmetry as ~ 6.2 subunits per turn of a ~ 250 Å pitch helix. All of the other layer lines in the power spectrum can be explained by this symmetry.
We used 3,678 overlapping segments for a three-dimensional reconstruction, employing the IHRSR method33. The reconstruction showed that the filament was clearly bipolar, that is, it was identical with itself when turned upside down. Since a single subunit of ICP8 cannot be bipolar, each asymmetric unit in the reconstruction must be a dimer of ICP8, and this was confirmed by STEM mass analysis.
STEM analysis of the mass per unit length of the ICP8 filaments
The mass per unit length of individual ICP8 filaments may be measured from dark-field STEM micrographs of unstained specimens (Fig. 2a) by integrating the corresponding image intensity36. The distribution of mass per unit length of the filaments was calculated relative to TMV particles as an internal standard (134.2 kDa/nm). The mass per length distribution measured (Fig. 3b) displays a single peak with the histogram fit very well with a Gaussian distribution. The average mass was 6.4 ± 0.3 kDa/Å (S.D.) (N=120). This would predict 12.5 subunits of ICP8 (each 128kDa) per 250 Å pitch helical turn, confirming the result from three-dimensional reconstruction that there are ~ 6.2 dimers of ICP8 per turn.
Fig. 2.
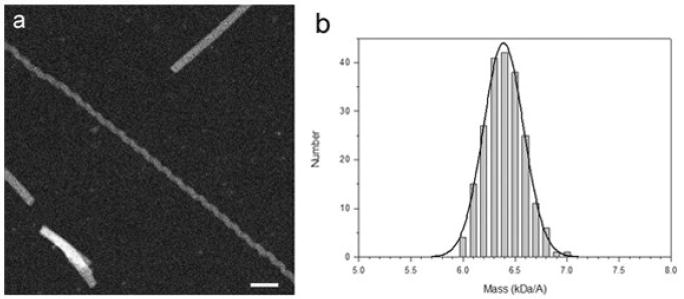
STEM microscopy and mass per unit length measurements of ICP8 filaments. (a) Dark-field micrograph of unstained freeze-dried specimens.Freshly prepared ICP8 filaments (50 – 100 μg/ml ICP8) were checked for optimal length and dispersal by negative staining. Freeze-dried specimens of the ICP8 filaments together with Tobacco mosaic virus (TMV) particles as a mass standard were prepared according to the standard “wet film” method of the Brookhaven STEM facility. This involved freeze-drying the sample with TMV on titanium grids supporting a 2-3 nm carbon film as described by Wall and Hainfeld37. Grids were scanned at -150° C on a custom-built STEM at 40 kV with a probe focused to 0.25 nm with a dwell time of 30 μs/pixel. Digital dark-field micrographs of freeze-dried specimens were recorded with 512 × 512 pixels at raster steps of 1.0 or 2.0 nm per pixel and an average radiation dose of 1 × 102 to 5 × 102 electron/nm2. The images were analyzed using the PCMass software (Brookhaven STEM resource). The resulting data were normalized to the known mass-per-unit-length of tobacco mosaic virus (131.4 kDa/nm). Histograms were calculated with 0.1 kDa/Å step. Scale bar equals 50 nm. (b) Histogram of distribution of ICP8 filament mass per unit length. A Gaussian distribution was fit to the histogram using the Marquardt-Levenberg algorithm (solid curve), as implemented in SigmaPlot (Systat Software). Average mass of ICP8 filaments was 6.4 ± 0.3 kDa/Å.
Fig. 3.
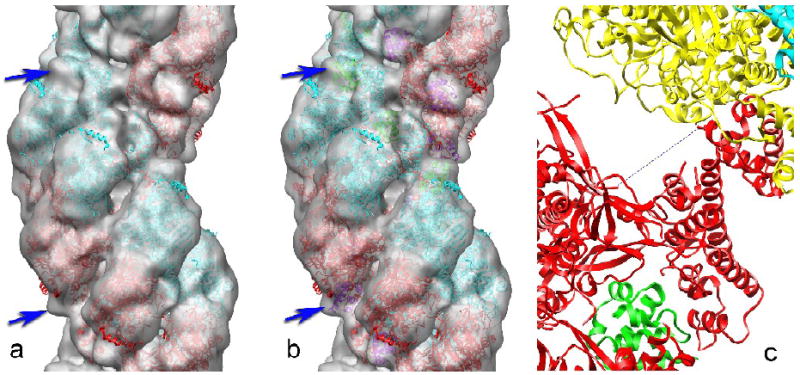
A three-dimensional reconstruction of the ICP8 filament is shown as a semi-transparent grey surface (a). Negatives were scanned using a Nikon Coolscan 8000 at 2000 dpi resulting in a final pixel size of 2.8 Å. Individual ICP8 filaments (n=181) were selected from several electron micrographs using the program ‘helixboxer’ within the EMAN software package34. We cut 3,678 overlapping segments of length 150 pixels from these filaments. The ICP8 filaments were reconstructed using the IHRSR (iterative helical real space reconstruction) method33, using a solid cylinder as the starting model. Reconstructions were done several times using different initial helical symmetries (the axial rise and rotation per subunit within the filament) to judge convergence. CHIMERA35 was used to fit the crystal structure of an ICP8 fragment into the three-dimensional reconstruction. The crystal structure of the large N-terminal domain of ICP82 has been fit into the reconstruction, with one copy in red and the symmetry-related copy in cyan. The dihedral symmetry present in these filaments requires that each asymmetric unit in the helix contains a symmetrical dimer of ICP8. This fit leaves a small volume in the reconstruction (blue arrows) empty of any atomic model. When the small C-terminal domain (residues 1048-1129) is placed into the reconstruction (b) in the same way that it is packed against a neighboring N-terminal domain in the ICP8 crystal2, this density becomes filled in the reconstruction (b, blue arrows). The C-terminal domain is shown in green for one strand, and in magenta for the opposite strand. The packing of a C-terminal domain from one subunit into a pocket within the N-terminal domain of an adjacent subunit is shown in (c). The C-terminal domain of the red subunit is packed against the N-terminal domain of the yellow subunit. The missing ten residues (dotted line) would need to span a distance of ~ 27 Å.
3-D reconstruction of ICP8 filaments and possible localization of the C-terminus
The dihedral symmetry (a two-fold axis perpendicular to the helical filament axis) was therefore imposed for subsequent cycles of IHRSR, but this introduced no major changes in the reconstructed volume (Fig. 3) other than reducing the noise. The surface of the reconstruction shown in Fig. 3a has been chosen to approximately enclose the expected molecular volume of ICP8. We have fit the reconstruction with an atomic model of ICP8 determined by x-ray crystallography2. One strand of subunits is shown in red (Fig. 3a), and the subunits related by the dyad symmetry are shown in cyan. The crystal structures could only be reasonably fit into the volume when it was reconstructed with a left-handed one-start helix, confirming the previous observation12 that these filaments are left-handed.
The crystal structure of ICP8 was from a C-terminal deletion, where 60 residues at the C-terminus were removed2. In addition, the last 7 residues in the construct used were not visualized in the crystallographic electron density map, so were not part of the atomic model generated. The crystal structure showed a very large N-terminal domain (the first 1038 amino acids), and a small C-terminal domain (residues 1049-1129) connected to the N-terminal domain by a flexible linker of ~ 10 residues that was not visualized due to disorder. The C-terminal domain of one subunit was observed to be separated from the N-terminal domain by 16-19 Å (there were two copies in the crystallographic asymmetric unit), and to be packed into a pocket formed by a neighboring N-terminal domain. A large rotation of the C-terminal domain with respect to the N-terminal domain existed between the two copies of ICP8 in the crystal, suggesting that the flexible linker placed few constraints on the orientation of the detached C-terminal domain. Since we had no reason to believe that the C-terminal domain of ICP8 would be bound in the pocket of a neighboring subunit as observed in the crystal, we simply truncated this domain and fit the large N-terminal domain into the reconstruction (Fig. 3a). The fit is quite reasonable, with only a single α-helix (residues 938-951) projecting out of the density envelope. This fit leaves a small volume (blue arrows, Fig. 3a) empty of an atomic model. We then added the crystal C-terminal domain (residues 1049-1129) to this model by placing it in the pocket of the neighboring N-terminal domain exactly as seen in the crystal (Fig. 3b). This model (Fig. 3b, blue arrows) now fills the density that was empty in Fig. 3a, suggesting that the crystal packing of the C-terminal domain into a neighboring N-terminal domain is maintained in the filament. The displacement of the C-terminal domain from the end of the N-terminal domain is now ~ 27 Å (Fig. 3c), which is more than observed in the crystal but still physically reasonable given the length of the flexible linker (~ 10 residues). Since we do not see any obvious density not explained by the atomic model, we suggest that the C-terminal 67 residues not included in the atomic model, but present in our filaments, are disordered. Consistent with this, the last 28 C-terminal ICP8 residues contain a nuclear localization signal38 that must be exposed to allow for recognition and nuclear import2.
A model for the location of ssDNA in the ICP8 filaments
Based upon structural similarity with other ssDNA binding proteins, Mapelli with co-authors proposed2 a model of the binding of ssDNA by ICP8. In this model, the ssDNA makes a rather sinuous path along a crystal chain of ICP molecules, necessary because the two copies of ICP8 in each asymmetric unit are related by a pseudo-21 screw axis. When the ssDNA bound to each subunit in this crystal model is applied to our filament, the ssDNA is now making an almost continuous path from one ICP8 subunit to the next (Fig. 4a), at a radius of ~ 25-30 Å from the helical axis. Because of the bipolar symmetry of these filaments, a second ssDNA molecule would run in the opposite direction, bound in an equivalent manner to the opposite strand of ICP8 subunits (Fig. 4b). While the proximity of two oppositely oriented single strands of DNA in such a filament is suggestive, we have no data at this point showing any such mechanism of annealing. We do know that the formation of dsDNA by ICP8 filaments causes these filaments to disassemble18, so we do not expect that there is a stable ICP8 filament containing the two single strands. It is of interest to note that the truncated version of ICP8 used for crystallography, ICP8ΔC, forms filaments on ssDNA10 which are very similar in appearance to the self-filaments of ICP8 that we have described in this paper. One can combine the observation from the previous study that the average axial rise per nucleotide was ~ 4.0 Å in those filaments with our observation that the axial rise per ICP8 subunit in these filaments is ~ 40 Å to arrive at a stoichiometry of ~ 10 nucleotides per ICP8 subunit. This is the same stoichiometry that was determined by solution studies using fluorescence anisotropy39.
Fig. 4.
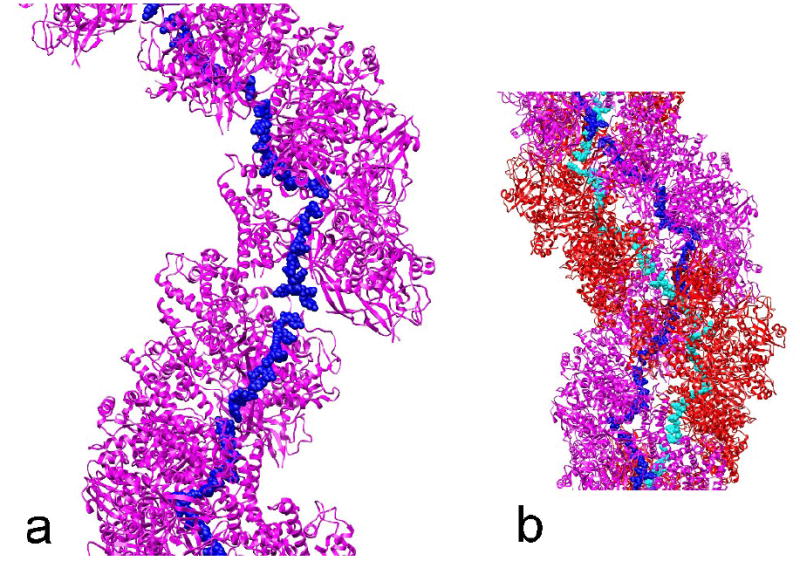
A combined model for the location of ssDNA in the ICP8 filament. The model for the binding of ssDNA by ICP8 as proposed previously2 has been applied to our filament model (Fig. 3b). A single strand of ICP8 subunits (magenta) is shown (a), and the ssDNA (blue atoms) bound to these ICP8 subunits would form an almost continuous strand at a helical radius of ~ 25-30 Å. When the second strand of ICP8 subunits (red), related by dihedral symmetry, is added to this model (b), the potential exists for a second strand of ssDNA (cyan atoms) to be bound. The second strand would run in the opposite orientation to the first strand, which would be the orientation present in dsDNA.
Acknowledgments
This work was supported by grants to E.H.E. (GM035269) and to J.D.G. (CA19014). Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu CA, Nelson NJ, McGeoch DJ, Challberg MD. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mapelli M, Panjikar S, Tucker PA. The crystal structure of the HSV-1 ssDNA binding protein suggests the structural basis for flexible, cooperative single-stranded DNA binding. J Biol Chem. 2005;280:2990–2997. doi: 10.1074/jbc.M406780200. [DOI] [PubMed] [Google Scholar]
- 3.Gupte SS, Olson JW, Ruyechan WT. The major herpes simplex virus type-1 DNA-binding protein is a zinc metalloprotein. J Biol Chem. 1991;266:11413–11416. [PubMed] [Google Scholar]
- 4.Powell KL, Littler E, Purifoy DJM. Non-structural proteins of herpes simplex virus. II. Major virus-specific DNA-binding protein. J Virol. 1981;39:894–902. doi: 10.1128/jvi.39.3.894-902.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, McNab MC, Perry LJ, Scott JE, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 6.Quinn JP, McGeoch DJ. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985;13:8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudas KC, Ruyechan WT. Identification of a region of the herpes simplex virus single-stranded DNA-binding protein involved in cooperative binding. J Virol. 1998;72:257–265. doi: 10.1128/jvi.72.1.257-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mapelli M, Muhleisen M, Persico G, van Der Zandt H, Tucker PA. The 60-residue C-terminal region of the single-stranded DNA binding protein of herpes simplex virus type 1 is required for cooperative DNA binding. J Virol. 2000;74:8812–8822. doi: 10.1128/jvi.74.19.8812-8822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruyechan WT. The major herpes simplex virus DNA-binding protein holds single-stranded DNA in an extended configuration. J Virol. 1983;46:661–666. doi: 10.1128/jvi.46.2.661-666.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mumtsidu E, Makhov AM, Konarev PV, Svergun DI, Griffith JD, Tucker PA. Structural features of the single-stranded DNA-binding protein of Epstein-Barr virus. J Struct Biol. 2008;161:172–187. doi: 10.1016/j.jsb.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 11.Ruyechan WT, Weir AC. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J Virol. 1984;52:727–733. doi: 10.1128/jvi.52.3.727-733.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Donnell ME, Elias P, Funnell BE, Lehman IR. Interaction between the DNA polymerase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J Biol Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 13.Lee SS, Lehman IR. Unwinding of the box I element of a herpes simplex virus type 1 origin by a complex of the viral origin binding protein, single-strand DNA binding protein, and single-stranded DNA. Proc Natl Acad Sci USA. 1997;94:2838–2842. doi: 10.1073/pnas.94.7.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, Lehman IR. Unwinding of a herpes simplex virus type 1 origin of replication (Ori(S)) by a complex of the viral origin binding protein and the single-stranded DNA binding protein. J Virol. 2000;74:5726–5728. doi: 10.1128/jvi.74.12.5726-5728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X, Lehman IR. An initial ATP-independent step in the unwinding of a herpes simplex virus type I origin of replication by a complex of the viral origin-binding protein and single-strand DNA-binding protein. Proc Natl Acad Sci USA. 2001;98:3024–3028. doi: 10.1073/pnas.061028298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makhov AM, Lee SS, Lehman IR, Griffith JD. Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: visualization of a specific preunwinding complex. Proc Natl Acad Sci USA. 2003;100:898–903. doi: 10.1073/pnas.0237171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutch RE, Lehman IR. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J Virol. 1993;67:6945–6949. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makhov AM, Griffith JD. Visualization of the annealing of complementary single-stranded DNA catalyzed by the herpes simplex virus type 1 ICP8 SSB/recombinase. J Mol Biol. 2006;355:911–922. doi: 10.1016/j.jmb.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 19.Bortner C, Hernandez TR, Lehman IR, Griffith JD. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J Mol Biol. 1993;231:241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 20.Nimonkar AV, Boehmer PE. The herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) promotes strand invasion. J Biol Chem. 2003;278:9678–9682. doi: 10.1074/jbc.m212555200. [DOI] [PubMed] [Google Scholar]
- 21.Nimonkar AV, Boehmer PE. On the mechanism of strand assimilation by the herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) Nucleic Acids Res. 2003;31:5275–5281. doi: 10.1093/nar/gkg740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reuven NB, Staire AE, Myers RS, Weller SK. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J Virol. 2003;77:7425–7433. doi: 10.1128/JVI.77.13.7425-7433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuven NB, Willcox S, Griffith JD, Weller SK. Catalysis of strand exchange by the HSV-1 UL12 and ICP8 proteins: potent ICP8 recombinase activity is revealed upon resection of dsDNA substrate by nuclease. J Mol Biol. 2004;342:57–71. doi: 10.1016/j.jmb.2004.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan MP, Chen LB, Knipe DM. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell. 1984;36:857–868. doi: 10.1016/0092-8674(84)90035-7. [DOI] [PubMed] [Google Scholar]
- 25.Zhou C, Knipe DM. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J Virol. 2002;76:5893–5904. doi: 10.1128/JVI.76.12.5893-5904.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S, Yu X, Seitz EM, Kowalczykowski SC, Egelman EH. Archaeal RadA protein binds DNA as both helical filaments and octameric rings. J Mol Biol. 2001;314:1077–1085. doi: 10.1006/jmbi.2000.5213. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 28.Makhov AM, Boehmer PE, Lehman IR, Griffith JD. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J Mol Biol. 1996;258:789–799. doi: 10.1006/jmbi.1996.0287. [DOI] [PubMed] [Google Scholar]
- 29.Stasiak A, Egelman EH. Structure and function of RecA-DNA complexes. Experientia. 1994;50:192–203. doi: 10.1007/BF01924002. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 31.Makhov AM, Taylor DW, Griffith JD. Two-dimensional crystallization of herpes simplex virus type 1 single-stranded DNA-binding protein, ICP8, on a lipid monolayer. Biochim Biophys Acta. 2004;1701:101–108. doi: 10.1016/j.bbapap.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Boehmer PE, Lehman IR. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 34.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Wall JS. Mass measurements with the electron microscope. In: Hren JJ, Goldstein JI, Joy DC, editors. Introduction to Analytical Electron Microscopy. Plenum Publishing Corp.; New York: 1979. pp. 333–342. [Google Scholar]
- 37.Wall JS, Hainfeld JF. Mass mapping with the scanning transmission electron microscope. Annu Rev Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 38.Gao M, Knipe DM. Distal protein sequences can affect the function of a nuclear localization signal. Mol Cell Biol. 1992;12:1330–1339. doi: 10.1128/mcb.12.3.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gourves AS, Le Tanguy GN, Villani G, Boehmer PE, Johnson NP. Equilibrium binding of single-stranded DNA with herpes simplex virus type I-coded single-stranded DNA-binding protein, ICP8. J Biol Chem. 2000;275:10864–10869. doi: 10.1074/jbc.275.15.10864. [DOI] [PubMed] [Google Scholar]


