Abstract
Chromosomal replication initiation requires the regulated formation of dynamic higher order complexes. Escherichia coli ATP-DnaA forms a specific multimer on oriC, resulting in DNA unwinding and DnaB helicase loading. DiaA, a DnaA-binding protein, directly stimulates the formation of ATP-DnaA multimers on oriC and ensures timely replication initiation. In this study, DnaA Phe-46 was identified as the crucial DiaA-binding site required for DiaA-stimulated ATP-DnaA assembly on oriC. Moreover, we show that DiaA stimulation requires only a subgroup of DnaA molecules binding to oriC, that DnaA Phe-46 is also important in the loading of DnaB helicase onto the oriC-DnaA complexes, and that this process also requires only a subgroup of DnaA molecules. Despite the use of only a DnaA subgroup, DiaA inhibited DnaB loading on oriC-DnaA complexes, suggesting that DiaA and DnaB bind to a common DnaA subgroup. A cellular factor can relieve the DiaA inhibition, allowing DnaB loading. Consistently, DnaA F46A caused retarded initiations in vivo in a DiaA-independent manner. It is therefore likely that DiaA dynamics are crucial in the regulated sequential progress of DnaA assembly and DnaB loading. We accordingly propose a model for dynamic structural changes of initial oriC complexes loading DiaA or DnaB helicase.
In many cellular organisms, multiple proteins form dynamic complexes on the chromosomal origin for the initiation of DNA replication. In Escherichia coli, ATP-DnaA forms a specific multimeric complex on the origin (oriC), resulting in an initiation complex that is competent in the replicational initiation (1–3). ATP-DnaA complexes, but not ADP-DnaA complexes, unwind the DNA duplex within the oriC DNA unwinding element (DUE)2 with the aid of superhelicity of oriC DNA and heat energy, resulting in the formation of open complexes (4, 5). At the unwound region, the loading of a DnaB replicative helicase is mediated by a DnaC helicase loader, resulting in the formation of the prepriming complex (6, 7). DnaG primase then complexes with DnaB loaded on the single-stranded (ss) region, which leads to primer synthesis and the loading of DNA polymerase III holoenzyme (8). The cellular ATP-DnaA level fluctuates during the replication cycle with a peak around the time of initiation (9). At the post-initiation stage, DnaA-ATP is hydrolyzed in a manner depending on ADP-Hda protein and the DNA-loaded form of the β-clamp subunit of the polymerase III holoenzyme, yielding inactive ADP-DnaA (10–13). This DnaA inactivation system is called RIDA (regulatory inactivation of DnaA). Hda consists of a short N-terminal region bearing a clamp-binding motif and a C-terminal AAA+ domain. This protein is activated by ADP binding, which allows interaction with ATP-DnaA in a DNA-loaded β-clamp-dependent manner. RIDA decreases the level of cellular ATP-DnaA in a replication-coordinated manner and represses extra initiation events (9–11).
The timing of chromosomal replication initiation is strictly regulated and needs to be linked to the regulation of the dynamic conformational changes in the DnaA-oriC complexes, as well as to the cellular ATP-DnaA levels. DiaA is a DnaA-binding protein that stimulates ATP-DnaA assembly on oriC and thus the initiation of replication (14, 15). DiaA mutants show delayed initiation and even asynchronous initiations of multiple origins when cells are rapidly growing and multiple rounds of replication are progressing simultaneously. DiaA is a homotetramer, and each protomer has a DnaA-binding site, which allows the simultaneous binding of multiple DnaA molecules to the homotetramer and the stimulation of cooperative binding of ATP-DnaA molecules on oriC.
DnaA consists of four functional domains as follows: the C-terminal domain IV has a DNA-binding helix-turn-helix structure (16) and domain III is an AAA+ domain that contains ATP-interacting motifs, homomultimer formation sites, and specific residues, termed B/H motifs, that can interact with ssDNA of the unwound DUE (17–21). Domain III forms a head-to-tail homomultimer whose overall structure is altered by ATP binding. It is possible that this multimer forms a spiral shape, in which one round of the spiral contains approximately seven protomers, and the resultant central pore carries the B/H motifs on the surface (21, 22). Domain II is a flexible, unstructured linker (23, 24), and domain I has a compactly folded structure, which interacts with several proteins including domain I per se, DiaA, and DnaB helicase (14, 15, 23, 25, 26). Domain I most likely forms homodimers in a head-to-head manner, which would line up the DnaB-interacting sites within this domain, thereby promoting DnaB loading (23).
E. coli oriC carries a dozen DnaA-binding sites, including the high affinity 9-mer DnaA boxes (R1 and R4 sites) and ATP-DnaA-preferential low affinity sites (ADLAS), which include the I and τ sites (20, 27). The interaction of ATP-DnaA with ADLAS is specifically important for the activation of DnaA-oriC complexes. DiaA stimulates the cooperative binding of ATP-DnaA on oriC, especially on ADLAS, resulting in the formation of open complexes (15). DnaB helicase stably complexes with DnaC, and the resulting DnaBC complexes can interact with open complexes, loading DnaB onto ssDNA of the unwound DUE. We have previously determined the tertiary structure of the DnaA domain I and found that DnaA Glu-21, within this domain, is a DnaB interaction site, specifically required for DnaB loading onto open complexes (23). The fundamental complex structure, the spatial organization of oriC-DnaA multimers complexed with DiaA, and those involved in the loading of DnaB onto oriC complexes have yet to be revealed.
In this study, our first step was the determination of a crucial DiaA-binding site, Phe-46, on DnaA domain I, using NMR and mutant analyses. Next we found that this site is required for DiaA-dependent stimulation of initiation complex formation and that only a subgroup of DnaA molecules, assembled on oriC, is sufficient for DiaA stimulation. Furthermore, we revealed that DnaA Phe-46 is also important for interactions with DnaB helicase. Like the DiaA stimulation, the stimulation of DnaB loading requires only a subgroup of DnaA molecules assembled on oriC. Competition analyses suggested that DiaA and DnaB interact with a common DnaA subgroup on oriC. Only a specific DnaA subgroup in an initiation complex might expose domain I to a position available for the protein loading. Cells might contain a modulator for the inhibition of DnaB loading by DiaA. Thus we infer that DiaA can regulate the initiation of replication both positively and negatively, i.e. it promotes ATP-DnaA assembly and inhibits DnaB loading, thereby ensuring the sequential and regulated progress of initiation reactions. In addition we propose a novel model for the structure of initiation complexes that includes DiaA and suggest possible modes of interactions for DiaA and DnaB on the initial complexes.
EXPERIMENTAL PROCEDURES
NMR Measurements and DiaA Titration
The NMR spectra were recorded at 25 °C on a Varian Unity INOVA 600 spectrometer. NMR samples of the 15N-labeled DnaA N terminus were dissolved in buffer (50 mm sodium phosphate (pH 6.5), 20 mm EDTA, 40 mm KCl, 2 mm dithiothreitol, and 10% sucrose containing 100% D2O or 90% H2O, 10% D2O). The chemical shift assignments for the DnaA N terminus in the same buffer were as described previously (28). DiaA P72A was prepared by triplicate dialysis against the same buffer. We recorded the 1H-15N HSQC spectra of 0.1 mm 15N-labeled DnaA in the presence of various concentrations (0, 10, 20, and 30 μm) of dialyzed DiaA. For the identification of the binding site between DiaA and DnaA, we recorded 1H-15N HSQC for 48 h at each DiaA concentration. The respective concentrations of the proteins were corrected for dilution.
Bacterial Strains, Media, Plasmids, and Purified Proteins
The E. coli K12 derivatives KH5402-1 (wild type), KA450 [ΔoriC1071::Tn10 dnaA17 (Am) rnhA199 (Am)], TK24 [dnaA204 diaA26::Tn5], KA451 [dnaA::Tn10 rnhA::cat], and YH105 [thyA rpsL ΔdiaA::FRT kan] have been described previously (15, 29, 30). The wild-type diaA in KA451 was replaced with ΔdiaA::FRT kan by P1 transduction using YH105, resulting in KK001. LB medium contained Bacto-tryptone (1%), yeast extract (0.5%), and NaCl (1%).
The plasmids pKA234, M13KEW101, and pOZ18 have also been described previously (29). For the construction of pDnaAF46A, a base substitution was introduced into the wild-type dnaA allele carried on pKA234 using a QuikChange site-directed mutagenesis kit (Stratagene). The mutagenic primers used for the construction of pDnaAF46A (bearing dnaA F46A) were 5′-GTACGCGCCAAACCGCGCGGTCCTTCGATTGGGTACG-3′, together with its complementary strand. For the construction of pRF46A, a 1.5-kb EcoRI fragment containing dnaA F46A was isolated from pDnaAF46A and inserted into the corresponding site of the low copy mini-R plasmid derivative, pOZ18. For the construction of pMZ1, a 5.7-kb BglII fragment was isolated from pOZ18, followed by self-ligation and exclusion of both dnaA and rnhA genes. For the construction of pRRNH, a 6.6-kb EcoRI fragment was isolated from pOZ18, followed by self-ligation and exclusion of the dnaA gene.
Hexahistidine-fused DiaA (His-DiaA), including wild-type DiaA, DiaA P72A, and DiaA F191L, was purified as described previously (15). Hexahistidine-fused DnaB (His-DnaB) was purified using a nickel - affinity purification system (31). Biotin-tagged DnaA (bio-DnaA) was purified as described previously (15).
Pulldown Assay for DiaA-DnaA Complexes
This assay was performed as described previously (15).
Pulldown Assay Using a Bio-oriC Fragment or Bio-DnaA
These assays were performed as described previously, with minor modifications (15). In an oriC pulldown assay, the indicated amounts of DnaA, His-DiaA, His-DnaB, DnaC, and biotin-tagged oriC fragment (419 bp) were incubated on ice for 15 min in buffer (10 μl) containing 50 mm HEPES-KOH (pH 7.6), 1 mm EDTA, 2 mm dithiothreitol, 20% sucrose, 100 mm KCl, 0.1 mg/ml bovine serum albumin, 1 mm ATP, 10 mm magnesium acetate, and 0.01% Triton X-100. The mixture was further incubated at 4 °C for 15 min with gentle rotation in the presence of streptavidin-coated beads (Promega) equilibrated in the same buffer (12.5 μl) as described above. The beads and bound material were collected, washed in buffer (12.5 μl) containing 50 mm HEPES-KOH (pH 7.6), 1 mm EDTA, 2 mm dithiothreitol, 20% sucrose, 50 mm KCl, 1 mm ATP, 10 mm magnesium acetate, and 0.01% Triton X-100, resuspended in 10 μl of SDS sample buffer, and analyzed by SDS-10−13% PAGE and silver staining. In a DnaA pulldown assay, the indicated amounts of DnaA, His-DiaA, an oriC fragment (419 bp), and bio-DnaA were incubated on ice for 15 min in the same buffer (10 μl) as above. bio-DnaA-bound materials were analyzed as above.
Reconstituted RIDA Reaction System
The staged RIDA reconstituted system was performed essentially as described previously (13, 23). Briefly, the DNA-loaded β-clamps were isolated using a gel filtration spin column as described previously (12, 13). Next, [α-32P]ATP-DnaA (0.25 pmol) was incubated at 30 °C for 20 min in buffer (12.5 μl) containing 20 mm Tris-HCl (pH 7.5), 8 mm dithiothreitol, 8 mm magnesium acetate, 0.01% Brij-58, 10% glycerol, 0.1 mg/ml bovine serum albumin, 120 mm potassium glutamate, 30 μm ADP, the indicated amounts of the C-terminally hexahistidine-fused Hda (Hda-cHis), and the DNA-loaded β-clamps. Nucleotides bound to DnaA were recovered on a nitrocellulose filter, separated by thin layer chromatography, and quantified by a BAS2500 bioimaging analyzer (Fuji Film).
DNase I Footprint Analysis
This analysis was performed essentially as described previously (15, 20, 21). Briefly, the indicated amounts of DnaA and His-DiaA were incubated at 30 °C for 10 min in buffer (10 μl) containing 25 mm HEPES-KOH (pH 7.6), 5 mm calcium acetate, 2.8 mm magnesium acetate, 35 mm ammonium sulfate, 23 mm potassium acetate, 4 mm dithiothreitol, 10% glycerol, 0.2% Triton X-100, 0.5 mg/ml bovine serum albumin, 14 μg/ml poly(dA-dT)-(dA-dT), 14 μg/ml poly(dI-dC)-(dI-dC), 3 mm ATP or ADP, and 5.5 ng of 32P-end-labeled oriC fragment (419 bp), which was amplified by PCR using a M13KEW101 oriC plasmid, followed by incubation at 30 °C for 4 min in the presence of DNase I (1.3 milliunits; Invitrogen). The resultant DNA samples were purified, separated by 5% sequence gel electrophoresis, and analyzed using a BAS2500 bioimaging analyzer (Fuji Film).
P1 Nuclease Assay for the Open Complex Formation
This assay was performed as described previously (15). The indicated amounts of DnaA were incubated at 38 °C for 3 min in buffer (50 μl) containing 60 mm HEPES-KOH (pH 7.6), 130 mm KCl, 0.1 mm zinc acetate, 8 mm magnesium acetate, 30% glycerol, 0.32 mg/ml bovine serum albumin, 5 mm ATP, 16 ng of HU protein, 320 ng of M13KEW101 RF I (61 fmol), and the indicated amounts of His-DiaA, followed by incubation at the same temperature for 200 s in the presence of P1 nuclease (10 units; Yamasa Co.). The resultant DNA samples were purified and digested with AlwNI, following by 1% agarose gel electrophoresis and ethidium bromide staining. The DNA fragments were quantified by densitometric scanning.
ABC Primosome Assay
This assay was performed as described previously (23, 29). Briefly, the indicated amounts of DnaA were incubated at 30 °C for 10 min in buffer (25 μl) containing M13-A site single-stranded DNA (220 pmol as nucleotide), 20 mm Tris-HCl (pH 7.5), 0.1 mg/ml bovine serum albumin, 8 mm dithiothreitol, 8 mm magnesium acetate, 0.01% Brij-58, 125 mm potassium glutamate, 0.5 μg of SSB, 117 ng of DnaB, 260 ng of DnaC, 72 ng of DnaG, 76 ng of DNA polymerase III*, 26 ng of β-clamp subunit, 1 mm ATP, 0.25 mm each of GTP, CTP, and UTP, 0.1 mm each of dNTP, and [α-32P]dTTP (50–100 cpm/pmol). DNA polymerase III* consists of the clamp loader complex and the DNA polymerase III core complex, which contains the catalytic center of the polymerase. [α-32P]dTTP was included to enable the subsequent measurement of DNA synthesis by liquid scintillation counting of acid-insoluble materials.
Form I* Formation Assay
This assay was performed essentially as described previously (23). Briefly, the indicated amounts of ATP-DnaA were incubated at 30 °C for 30 min in buffer (12.5 μl) containing 20 mm Tris-HCl (pH 7.5), 0.1 mg/ml bovine serum albumin, 8 mm dithiothreitol, 10 mm magnesium acetate, 125 mm potassium glutamate, 2 mm ATP, 1.2 μg of SSB, 1.3 ng of HU protein, 68 ng of DnaB, 46 ng of DnaC, 90 ng of DNA gyrase A subunit, 113 ng of DNA gyrase B subunit, and 100 ng of M13KEW101 RF I (19 fmol). The reaction was stopped in the presence of SDS and EDTA, followed by 0.65% agarose gel electrophoresis and ethidium bromide staining. The produced form I* DNA was quantified by densitometry.
In Vitro Minichromosome Replication System
This system was performed essentially as described previously (14, 15). Briefly, the indicated amounts of ATP-DnaA and His-DiaA were incubated at 30 °C for 20 min in buffer (25 μl) containing 40 mm HEPES-KOH (pH 7.6), 40 mm phosphocreatine, 2 mm ATP, 0.5 mm each of GTP, CTP, and UTP, 10 mm magnesium acetate, 0.1 mg/ml creatine kinase, and 7% polyvinyl alcohol, 240 μg of crude protein extract (TK24 fraction II), 200 ng of M13KEW101 RF I (600 pmol as nucleotide), 0.1 mm each of dNTP, and [α-32P]dTTP (50–100 cpm/pmol).
Flow Cytometry Analysis
This analysis was performed essentially as described previously (15, 29). Briefly, cells were grown in LB medium containing thymine (50 μg/ml) and ampicillin (50 μg/ml) exponentially for 10 generations at 30 °C until the absorbance (A660) reached 0.2. Incubation was further continued at the same temperature for 4 h in the presence of rifampicin (300 μg/ml) and cephalexin (10 μg/ml). Cells were fixed in cold 70% ethanol, washed, and stained with 2 μm SYTOX green (Invitrogen), followed by analysis using a FACSCalibur flow cytometry (BD Biosciences).
Immunoblot Analysis
This analysis was performed as described previously (15, 29).
RESULTS
Structural Model of a DiaA-DnaA Domain I Complex
Previously we determined the structures of the DiaA homotetramer, using x-ray analysis, and the DnaA N-terminal domains I and II (residues 1–108), using NMR analysis (15, 23). In addition, we showed that DiaA binds directly to DnaA domain I (residues 1–86) (14, 15). Based on these results, we performed NMR analysis using DiaA and 15N-labeled DnaA domains I and II, to determine the mode of the DiaA-DnaA domain I interaction. When we used wild-type DiaA, severe signal broadening occurred, and site-specific chemical shifts were not detected, probably because of the increased molecular size because of tight binding between the two proteins (data not shown).
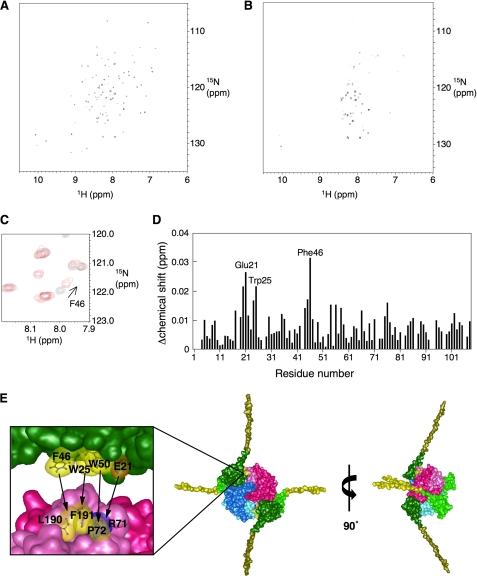
When we used DiaA P72A, which has a considerably reduced specific affinity for DnaA (15), specific chemical shifts were observed. As the concentration of DiaA P72A increased, signal broadening occurred in the 1H-15N HSQC spectra of the N terminus of the DnaA protein, especially in domain I (Fig. 1, A and B), consistent with an interaction between DiaA and DnaA domain I. When the molar ratio of DnaA:DiaA was 1:0.2, we found slight, but significant, chemical shift changes for several specific residues, including, in particular, Glu-21, Trp-25, and Phe-46 (Fig. 1, C and D).
FIGURE 1.
NMR analysis and the structural model of the DiaA-DnaA N-terminal complex. A–D, NMR analysis. The 1H-15N HSQC spectrum of the DnaA N terminus was obtained in the absence (A) and presence (B) of 20 μm DiaA P72A. Part of the 1H-15N HSQC spectrum, in the absence (black) and presence (red), of 20 μm DiaA P72A shows the chemical shift change for DnaA Phe-46 (C). DiaA-dependent chemical shift changes are displayed for the backbone amides as a function of the amino acid residue number (D). All chemical shifts changes in the 1H-15N HSQC spectra were calculated according to the formula (Δδ(1H)2 + (Δδ(15N)/7)2)1/2. Glu-21, Trp-25, and Phe-46 are indicated on the graph. E, structural model for the DiaA-DnaA N-terminal complex illustrated using MolFeat (FiatLux). This model is based on the structures of a DiaA homotetramer (15) and the DnaA N terminus (23). Each protomer of the DiaA tetramer is depicted in different colors. Domain I (residues 1–86) and part of domain II (residues 87–108), within the DnaA N terminus, are colored in green and ocher, respectively. The hydrophobic residues of DiaA Pro-71, Leu-190, and Phe-191 or DnaA Trp-25, Phe-46, and Trp-50 are colored in yellow. The charged residues of DiaA Arg-71 and DnaA Glu-21 are colored in deep blue and orange, respectively. The arrows show the residues with mutual affinity between DiaA and DnaA domain I.
Based on the results above, we constructed a structural model of the DiaA-DnaA domain I complex (Fig. 1E). DnaA Glu-21 and Phe-46 are exposed on the protein surface where they form a small patch together with Trp-25 and Trp-50 (23) (Fig. 1E). Our previous study revealed that DiaA residues Arg-71, Pro-72, Leu-190, and Phe-191 have specific roles in DnaA binding and that these residues form a patch on the DiaA protein surface (15) (Fig. 1E). Thus it is reasonable to infer that this patch on DiaA is used for direct binding to the patch on DnaA, which includes Glu-21 and Phe-46.
In this model, DnaA Phe-46 and Trp-25 are associated with DiaA Leu-190 and Phe-191 through hydrophobic interaction, consistent with the NMR data (Fig. 1D). This interaction most likely plays a crucial role in the formation of a stable DnaA-DiaA complex. Unlike DiaA L190A/L190P and DiaA F191L/F191S, DiaA R71Q and DiaA P72A/P72S have a certain level of residual DnaA binding activity (15). DnaA Glu-21 and Trp-50 would interact with DiaA Arg-71 and Pro-72, respectively, through ionic bonds and hydrophobic interaction, assisting in complex formation (Fig. 1E). Moreover, this arrangement explains the subtle chemical shift change of DnaA Trp-50, observed in the present NMR analysis using DiaA P72A. DnaA Glu-21 and Trp-25 reside on a short α-helix, and Thr-20 resides on its flanking loop (23), suggesting that the chemical shift change in Thr-20 is caused indirectly by a conformational change in Glu-21 (Fig. 1D). Moreover, this model is consistent with our previous data, which showed that a DiaA tetramer can bind multiple DnaA molecules (15). This new structural model coincides overall with the NMR and biochemical data for DiaA-DnaA complex formation and was thus adopted as the basis for further analysis. Indeed, this model is also consistent with the DiaA binding activities of DnaA mutants (see below).
DnaA F46A Mutant Fails to Bind DiaA
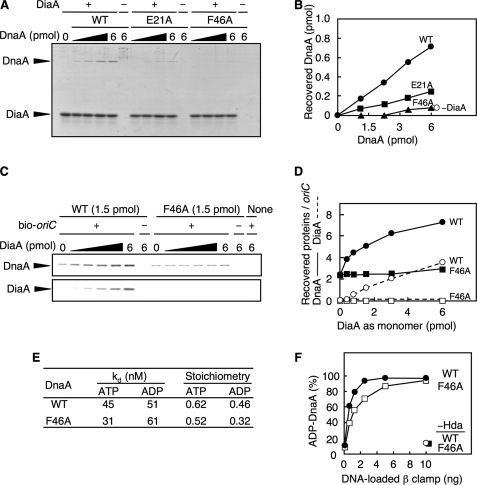
To analyze the specific roles of DnaA Phe-46 and Glu-21 in DiaA binding, DnaA F46A was overproduced and purified using our method for wild-type DnaA and DnaA E21A, as described previously (23) (supplemental Fig. S1A). We performed a pulldown assay using the DnaA samples and hexahistidine-fused DiaA (His-DiaA). Unlike wild-type DnaA, the DiaA binding activity of DnaA F46A was severely impaired (Fig. 2, A and B). The DiaA binding of DnaA E21A was also impaired, although it retained ∼30% residual activity. These results coincided well with our NMR analysis and the model for DiaA-DnaA domain I interaction (Fig. 1). As DnaA Phe-46 plays a prominent role in DiaA binding, it was mainly used in the following analysis.
FIGURE 2.
DnaA F46A is specifically defective in DiaA binding. A and B, indicated amounts of wild-type DnaA (WT), DnaA E21A, and F46A were incubated on ice for 5 min in the presence (+) or absence (−) of His-DiaA (5 pmol as monomer). Protein complexes were isolated using Co2+-conjugated beads, eluted in 1% SDS, and analyzed by SDS-12% PAGE and Coomassie Brilliant Blue staining (A). The DnaA bands were quantified by densitometry, and the recovered amounts were determined using a standard curve (B). C and D, indicated amounts of His-DiaA (as monomer) were incubated on ice for 15 min in the presence (+) or absence (−) of a biotin-tagged oriC fragment (bio-oriC; 100 fmol) and/or DnaA (1.5 pmol) of the wild-type form or the F46A mutant. Proteins bound to bio-oriC were isolated using streptavidin-beads (C). The protein bands were quantified by densitometry, and the recovered amounts of DnaA and DiaA were determined using standard curves (D). E, affinities of DnaA for ATP or ADP were determined using DnaA (2 pmol) and a filter-retention assay (29). Kd and stoichiometry were calculated by Scatchard plot. F, DnaA-ATP hydrolysis was assessed using a staged RIDA-reconstituted system. The [α-32P]ATP-DnaA (0.25 pmol) was incubated at 30 °C for 20 min in buffer containing the indicated amounts of the DNA-loaded clamp in the presence, or absence, of Hda protein (45 fmol). The ratio of ADP-DnaA to total ATP-/ADP-DnaA is shown as a percentage.
Wild-type DiaA stimulates DnaA assembly on oriC by linking multiple DnaA molecules and enhancing cooperative binding (15). When this function was assessed for the presence of DnaA F46A by a pulldown assay, using biotinylated-oriC DNA (419 bp) as described previously (15), the assembly of DnaA F46A on oriC was not stimulated by DiaA, and DiaA was not recovered with the oriC-DnaA complexes (Fig. 2, C and D, and supplemental Fig. S1B), consistent with the above results (Fig. 2, A and B). Under the same conditions, the recovery of the wild-type DnaA increased in a DiaA-dependent manner as we reported previously (15) (Fig. 2, C and D, and supplemental Fig. S1B). A gel mobility shift assay indicated that the activity of DnaA F46A in binding to a single DnaA box was comparable with that of the wild-type DnaA (supplemental Fig. S1, C and D), consistent with the data from the oriC-pulldown experiments in the absence of DiaA (Fig. 2, C and D) and with the idea that DnaA F46A, specifically, is defective in DiaA-dependent DnaA assembly on oriC.
DnaA F46A clearly has the same affinity for ATP and ADP as wild-type DnaA (Fig. 2E and supplemental Fig. S1, E and F). In a RIDA reaction, DnaA-ATP was efficiently hydrolyzed in a manner depending on Hda and the DNA-loaded β-clamp, which is required for the replication cycle-coordinated fluctuation of the cellular ATP-DnaA level (10–12). The DnaA domain I is required for RIDA-specific DnaA-ATP hydrolysis (12). When analyzed using a reconstituted RIDA system, DnaA F46A was basically active during RIDA-specific DnaA-ATP hydrolysis compared with wild-type DnaA (Fig. 2F and supplemental Fig. S1G). These results support the idea that DnaA Phe-46 plays a specific role in the interaction with DiaA.
DnaA F46A Can Form ATP-DnaA-specific oriC Complexes but Is Insensitive to DiaA
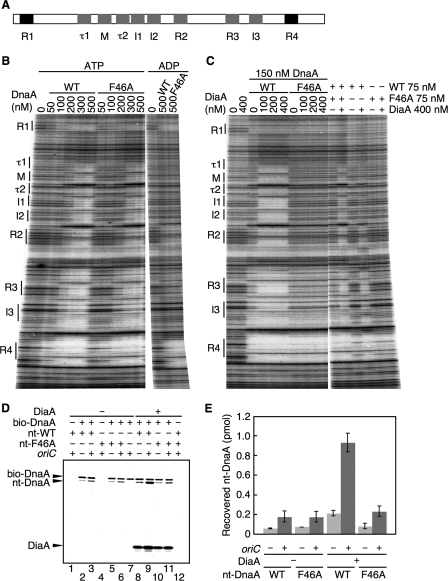
Unlike ADP-DnaA, ATP-DnaA efficiently forms multimers complexed with ADLAS on oriC (20, 27) (Fig. 3A). When assessed by a DNase I footprint assay using an oriC fragment as we reported previously (20, 21), ATP-DnaA F46A and wild-type ATP-DnaA exhibited similar patterns (Fig. 3B). At a DnaA concentration of 100 nm, both wild-type DnaA and DnaA F46A bound to the high affinity DnaA boxes, R1 and R4. In addition, these proteins associated with ADLAS at a DnaA concentration of 200 nm (for R2, R3, and I3) and at a DnaA concentration of 300 nm (for τ1, M, τ2, I1, and I2), in an ATP form-specific manner (Fig. 3A). These results indicate that basically DnaA F46A retains the ability to form ATP-DnaA specific complexes on oriC. This can be explained by the presence of the intact domains III and IV in DnaA F46A.
FIGURE 3.
DnaA F46A is defective in DiaA-dependent formation of DnaA-oriC complexes. A, structure of oriC DnaA-binding region. The positions of the DnaA-binding sequences are indicated. High affinity sites (R1 and R4) and ADLAS (R2, R3, M, and I/τ sites) are colored in black and gray, respectively. B, amounts of wild-type DnaA (WT) and DnaA F46A (F46A) indicated were incubated at 30 °C for 10 min in buffer containing a 32P-end-labeled oriC fragment and 3 mm ATP or ADP, followed by DNase I footprint analysis using 5% sequencing gel electrophoresis. The positions of the DnaA-binding sequences (boxes R1–R4 and M; sites I1–3 and τ1–2) are indicated. C, indicated amounts of DiaA (as monomers) were incubated at 30 °C for 10 min, in the presence of 3 mm ATP, with the indicated amounts of wild-type DnaA and/or DnaA F46A, followed by DNase I footprint analysis as described for B. D, reactions were incubated on ice for 15 min in the presence (+) or absence (−) of His-DiaA (500 nm; 5 pmol as monomer), bio-DnaA (250 nm; 2.5 pmol), nontagged wild-type DnaA (nt-WT) (250 nm; 2.5 pmol), DnaA F46A (nt-F46A) (250 nm; 2.5 pmol), oriC fragment (10 nm; 100 fmol). Proteins bound to streptavidin beads were isolated and analyzed by SDS-10% PAGE and silver staining. Nt-DnaA, nontagged DnaA. E, recovered amounts of nt-WT and nt-F46A were determined using standard curves, and the average values from two independent experiments were indicated.
DiaA promotes the formation of ATP-DnaA-specific oriC complexes, even when the level of DnaA was limited (15). Therefore, next we performed similar analyses using 150 nm DnaA in the presence, or absence, of DiaA (Fig. 3C). In the absence of DiaA, wild-type ATP-DnaA and ATP-DnaA F46A interacted with the DnaA box R1 and with a region that included the DnaA boxes R3/R4 and the I3 site (Fig. 3C). The DnaA box R3 and the I3 site reside close to the high affinity box R4, which most likely assists in the cooperative binding of DnaA at this DnaA concentration. This idea is also consistent with in vivo results of oriC mutant analysis (32). Also, at this concentration, it was just possible to detect binding to the DnaA box R2 (Fig. 3C), consistent with previous reports (20, 21). In the presence of DiaA, wild-type ATP-DnaA also interacted with ADLAS from the τ1 to I2 sites, but ATP-DnaA F46A did not (Fig. 3C). Together with the data from the oriC pulldown experiments (Fig. 2, C and D, and supplemental Fig. S1B), these results demonstrate a crucial role for DnaA Phe-46 in the DiaA-dependent efficient formation of initiation complexes.
DiaA-dependent ATP-DnaA Complex Formation Requires Only a Subgroup of DnaA Molecules on oriC
To elucidate further the structure of ATP-DnaA-oriC complexes containing DiaA, we analyzed mixtures of the ATP forms of wild-type DnaA and DnaA F46A. When we used a 1:1 mixture of wild-type ATP-DnaA and ATP-DnaA F46A at a total DnaA concentration of 150 nm, the resultant footprint patterns, in the absence (and in the presence) of DiaA, were similar to those obtained for wild-type ATP-DnaA at 150 nm (Fig. 3C), and protection at ADLAS from τ1 to I2 was clearly detected in a DiaA-dependent manner. In contrast, when only wild-type ATP-DnaA was included at 75 nm, the protection of τ1, M, and I2 sites was not detected, and τ2 and I1 sites were only slightly protected even in the presence of DiaA. These results can be consistent with the idea that only a subgroup of DnaA molecules, assembled on oriC, interacts with DiaA, thereby enhancing the formation of ATP-DnaA-specific oriC complexes.
In the above experiments, we assumed that a mixed multimer of wild-type DnaA and DnaA F46A was formed on oriC. To directly elucidate this assumption, we performed a pulldown assay using biotin-tagged and nontagged DnaA proteins. Nontagged wild-type DnaA or DnaA F46A was incubated in the presence or absence of bio-DnaA, DiaA, and an oriC fragment. Materials bound to bio-DnaA were collected using streptavidin beads. The recovery of nontagged wild-type DnaA and DnaA F46A increased 2–3-fold in an oriC-dependent manner in the absence of DiaA (Fig. 3, D, lanes 2, 3, 5, 6, and E). Nontagged DnaA was slightly recovered even in the absence of oriC, which would be caused by weak inter-DnaA interaction. Similar results were obtained even in the presence of DiaA (Fig. 3D, lanes 8–11). DiaA-dependent overall increase in the recovery of nontagged wild-type DnaA (Fig. 3D, lanes 8 and 9) is consistent with previous data. Nontagged DnaA and DiaA did not directly bind to streptavidin beads (Fig. 3D, lanes 1, 4, 7, and 12). These results indicate that DnaA F46A as well as wild-type DnaA can form a mixed multimer with biotin-tagged wild-type DnaA (bio-DnaA) on the oriC fragment.
DiaA-dependent Stimulation of oriC Unwinding Activity
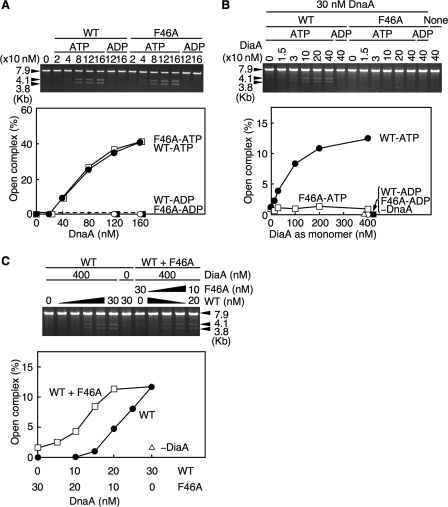
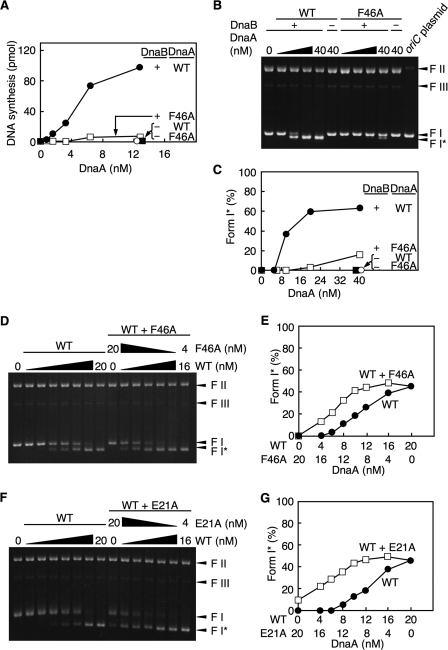
To characterize the role of DnaA F46A during initiation at oriC, we assessed open complex formation using an oriC plasmid and P1 nuclease, which specifically cleaves ssDNA. If the oriC DUE on the plasmid is cleaved by P1 nuclease, subsequent digestion with AlwNI restriction enzyme will produce two fragments of 3.8 and 4.1 kb (21). As for wild-type ATP-DnaA, ATP-DnaA F46A, but not ADP-DnaA F46A, was active in open complex formation (Fig. 4A). When limited amounts of wild-type DnaA were included, DiaA promoted open complex formation in an ATP-DnaA-dependent manner (Fig. 4B), consistent with our previous report (15). In contrast, DnaA F46A was not stimulated during open complex formation by DiaA (Fig. 4B), consistent with the footprint analysis (Fig. 3C) and the idea that DiaA-dependent stimulation of open complex formation requires DnaA Phe-46.
FIGURE 4.
DnaA F46A activities in open complex formation. A, amounts of the ATP or ADP form of the wild-type DnaA (WT) and DnaA F46A (F46A) indicated were incubated at 38 °C for 3 min in buffer containing M13KEW101 oriC plasmid and HU protein, followed by further incubation with P1 nuclease. The resultant DNA was purified, digested with AlwNI, and analyzed using 1% agarose gel electrophoresis and ethidium bromide staining. The intensities of the band signals were quantified by densitometry. The total amounts of the 3.8- and 4.1-kb fragments were normalized to those of the total DNA and were plotted as an open complex (%). B, indicated amounts of DiaA (as a monomer) were similarly incubated in the presence or absence of the ATP or ADP form of wild-type DnaA or DnaA F46A (30 nm; 1.5 pmol). C, reaction mixtures containing the indicated amounts of wild-type DnaA and DnaA F46A, or of the wild-type DnaA alone, were similarly analyzed in the presence or absence of DiaA (400 nm; 20 pmol as a monomer).
Next we analyzed whether DiaA enhances the open complex formation of a hetero-mixture of DnaA F46A and the wild-type DnaA. When mixtures of DnaA F46A and wild-type DnaA were used, open complex formation was enhanced in a DiaA-dependent manner compared with that of the wild-type DnaA alone (Fig. 4C). Notably, when a mixture of 20 nm wild-type DnaA and 10 nm DnaA F46A was used, the DiaA-dependent unwinding activity was elevated to that equivalent to 30 nm of the wild-type DnaA alone (Fig. 4C). These results were consistent with those from the footprint experiments using a similar mixture (Fig. 3C). Thus, these results further support the idea that only a subgroup of DnaA molecules assembled on oriC is needed to interact with DiaA and to stimulate initiation.
DnaA Phe-46 Is Also Important for DnaB Helicase Loading
In the initiation process, DnaB helicase is loaded onto the replication origin after open complex formation. We previously reported that the DnaA E21A mutant protein is specifically defective in DnaB helicase loading (23). This mutant protein also exhibited reduced DiaA binding activity (Fig. 2A), which suggests that the patch, including Glu-21 on DnaA domain I, could function for both DiaA binding and DnaB loading (Fig. 1E). We thus investigated the role of DnaA F46A in the DnaB loading process.
First we used the ABC primosome system for ssDNA replication (33). In this system, the template is ssDNA with a local hairpin structure containing a DnaA box. DnaA bound to this hairpin directs DnaB loading onto the ssDNA, which enables replication by DnaG primase and DNA polymerase III holoenzyme. DnaA F46A was inactive in this reaction, suggesting that DnaA F46A is defective in DnaB loading (Fig. 5A).
FIGURE 5.
DnaB helicase loading activities of DnaA F46A. A, ABC primosome assay. The indicated amounts of the wild-type DnaA (WT) and DnaA F46A (F46A) were incubated at 30 °C for 10 min in buffer containing DnaC helicase loader, SSB, DnaG primase, DNA polymerase III holoenzyme, and M13 A-site ssDNA (220 pmol as nucleotide) in the presence (+) or absence (−) of DnaB helicase. B–G, form I* assay. The indicated amounts of the wild-type DnaA (WT) and DnaA F46A were incubated at 30 °C for 30 min in buffer containing DnaC, SSB, HU protein, DNA gyrase, and M13KEW101 oriC plasmid in the presence (+) or absence (−) of DnaB helicase. DNA was purified and analyzed using 0.65% agarose gel electrophoresis and ethidium bromide staining. The migration positions of form I (FI; supercoiled form), form I* (FI*; hyper-supercoiled form), form II (FII; relaxed form), and form III (FIII; linear form) are indicated. The input form of M13KEW101 was also analyzed (oriC plasmid) (B). The intensities of the band signals were quantified, and the amounts of form I* DNA are shown as percentages of the total DNA (C). D and E, reaction mixtures containing the indicated amounts of wild-type DnaA and DnaA F46A, or wild-type DnaA alone, were analyzed as described above. F and G, similar analysis to that described above was performed using DnaA E21A instead of DnaA F46A.
We then investigated the role of DnaA F46A in DnaB helicase loading onto oriC using a supercoiled (form I) oriC plasmid and a form I* assay. After unwinding of oriC, DnaA recruits the DnaB-DnaC complex via a direct interaction with DnaB helicase, resulting in DnaB loading onto the unwound region and DnaC dissociation. DnaB helicase further unwinds the duplex DNA in the presence of DNA gyrase, resulting in production of a highly negative supercoiled form called form I*. The form I* oriC plasmid can be separated from form I oriC plasmid by agarose gel electrophoresis (34). Unlike wild-type DnaA, DnaA F46A had severely impaired activity in this assay (Fig. 5, B and C). A slight residual activity was seen by excessive supply of DnaA F46A. These results indicate that DnaA Phe-46 plays an important role in DnaB helicase loading and thus in prepriming complex formation.
DnaB Helicase Loading on oriC Requires Only a Subgroup of DnaA Molecules
To determine whether all DnaA molecules included in an open complex are required for DnaB helicase loading, we used form I* assay mixtures, including wild-type DnaA and DnaA F46A or DnaA E21A, at various ratios (Fig. 5, D–G). When we used a mixture including wild-type DnaA and DnaA F46A in a 3:2 ratio, the activity of the DnaB helicase loading reached a level corresponding to the maximum of the wild-type DnaA alone (Fig. 5, D and E). Similar results were obtained for DnaA E21A (Fig. 5, F and G). These results suggest that the role played by the DnaA domain I in DnaB loading is performed by only a subgroup of DnaA molecules assembled on oriC. When mixtures of DnaA E21A and F46A were similarly used, DnaB loading activity was not observed (data not shown).
In the above experiments, based on the pulldown assay (Fig. 3, D and E), we assumed that a mixed multimer of wild-type DnaA and DnaA F46A is formed even on oriC plasmid used for P1 assay and form I* assay. However, we do not exclude implausible possibilities, e.g. DnaA F46A could indirectly activate initiation complexes, including only wild-type DnaA on oriC plasmid. Alternatively, only a subset, but not a majority, of oriC complexes, including both wild-type DnaA and DnaA F46A, could have an abnormally high activity in P1 assay and form I* assay.
DnaB Helicase Forms a Stable Complex with DnaA Molecules on oriC
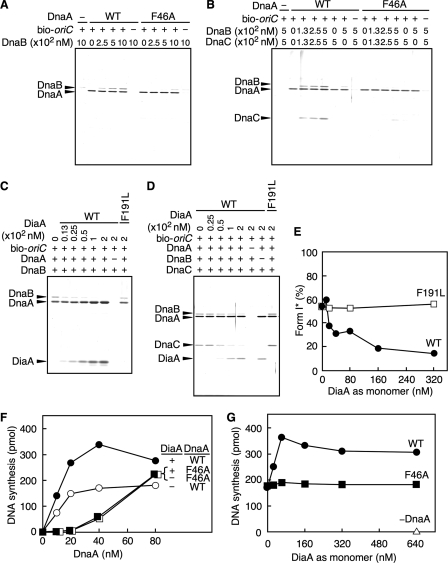
To analyze the mechanics of the interaction between DnaB helicase and the DnaA multimers on oriC, we performed an oriC pulldown assay using DnaA, DnaB, and DnaC. We also used hexahistidine-fused DnaB (His-DnaB) to distinguish DnaA from DnaB by SDS-PAGE. The activity of His-DnaB was similar to that of the native DnaB in a form I* assay (data not shown). In the absence of oriC, complexes formed only by DnaA and DnaB were not been detected by pulldown assay because of a weak affinity between the two proteins. However, in the present assay, DnaB helicase was observed to complex with DnaA multimers bound to oriC in the absence, and presence, of a DnaC helicase loader (Fig. 6, A and B). This is most likely because the multimeric DnaA molecules on oriC provide a DnaB-homohexamer with multiple, and regularly aligned, DnaB-binding sites, thereby resulting in an increase in the overall affinity. This idea is consistent with our structural model (23) and the fact that DnaB helicase loading onto open complexes requires stable multimers of DnaA (19). Unlike wild-type DnaA, the binding of DnaA F46A to DnaB was severely impaired even in the presence of oriC (Fig. 6, A and B), consistent with the data from the form I* assay (Fig. 5, B and C). These results support the idea that the DnaB helicase can load onto initiation complexes as well as onto open complexes, and that DnaA Phe-46 is important for DnaB helicase loading on these complexes.
FIGURE 6.
Loading of DiaA and DnaB helicase to DnaA complexes on oriC. A, indicated amounts of His-DnaB (as monomer) were incubated for 15 min on ice in the presence (+) or absence (−) of bio-oriC (10 nm; 100 fmol) and 500 nm (5 pmol) of wild-type DnaA (WT) or DnaA F46A. Proteins bound to bio-oriC were isolated using streptavidin-beads and analyzed. B, indicated amounts of His-DnaB (as monomer) and DnaC were incubated, as above, in the presence (+) or absence (−) of DnaA (500 nm; 5 pmol) and bio-oriC (10 nm; 100 fmol) and were analyzed as described above. C, indicated amounts of wild-type DiaA (WT) and DiaA F191L (as monomer) were incubated, as above, in buffer containing DnaB (500 nm; 5 pmol as monomer) and bio-oriC (10 nm; 100 fmol) in the presence (+) or absence (−) of DnaA (500 nm; 5 pmol). Proteins bound to bio-oriC were analyzed as described above. D, indicated amounts of wild-type DiaA (WT) and DiaA F191L (as monomer) were incubated, as above, in buffer containing DnaC (500 nm; 5 pmol) and bio-oriC (10 nm; 100 fmol) in the presence (+) or absence (−) of DnaA (500 nm; 5 pmol) and His-DnaB (500 nm; 5 pmol as monomer). Proteins bound to bio-oriC were analyzed as described above. E, form I* assay. ATP-DnaA (24 nm; 0.3 pmol) and the indicated amounts of wild-type DiaA (WT) and DiaA F191L (as monomer) were used for this assay. The resultant DNA was analyzed as described for Fig. 5, B–G. F and G, indicated amounts of the ATP form of wild-type DnaA (WT) and DnaA F46A were incubated at 30 °C for 20 min in buffer containing M13KEW101 oriC plasmid (600 pmol as nucleotide) in the presence (+) or absence (−) of DiaA (160 nm; 4 pmol as monomer) using a TK24-derived crude replicative extract (F). The indicated amounts of DiaA (as monomer) were similarly analyzed using the same extract as that used in F in the presence (60 nm; 1.5 pmol) or absence (−) of wild-type DnaA (WT) or DnaA F46A (G).
Furthermore, in this assay, we found that the DnaC helicase loader was recovered in a DnaB-dependent manner and promoted the co-recovery of DnaB with DnaA-oriC complexes (Fig. 6B). It is possible that DnaC binds weakly but directly to DnaA complexed on oriC, thus assisting in stable binding between DnaA and DnaB. In other words, DnaA-DnaB-DnaC complexes on oriC might be further stabilized via DnaA-DnaC interaction, in addition to DnaA-DnaB and DnaB-DnaC interactions.
DiaA and DnaB Helicase Compete in Associating with DnaA Multimers on oriC
As both DiaA and DnaB commonly use DnaA Phe-46 for interactions, we used the oriC pulldown assay to analyze the correlation between DiaA and DnaB in interactions with DnaA multimers formed on oriC. First, we observed that the binding of DnaB to the DiaA-DnaA-oriC complexes was severely inhibited in a DiaA-dependent manner (Fig. 6C). This inhibition was also seen even in the presence of DnaC (Fig. 6D). In contrast, DnaB binding was not inhibited when the assay used DiaA F191L, which is specifically inactive in DnaA binding (15) (Fig. 6, C and D), suggesting that specific DiaA-DnaA binding inhibits DnaB loading. Consistently, DiaA-dependent inhibition was also seen in the form I* assay (Fig. 6E) and in a reconstituted replication system of minichromosomes using purified replicative proteins (data not shown). Taken together, these results support the ideas that DiaA basically precedes DnaB in binding to DnaA-oriC complexes and that DiaA and DnaB helicase bind directly to a common subgroup of DnaA molecules on oriC.
Replication Activity of DnaA F46A Is Impaired
Next we analyzed the initiation activity of DnaA F46A using a crude replication system. When we used a crude replicative fraction prepared from TK24 cells [dnaA204 diaA26::Tn5], wild-type DnaA promoted minichromosomal replication, which was stimulated 2-fold by DiaA (Fig. 6, F and G), consistent with our previous data (14, 15). In contrast, the activity of DnaA F46A was severely inhibited compared with that of wild-type DnaA, and this inhibition was not affected by DiaA (Fig. 6F). This result is consistent with the defects of DnaA F46A in DnaB loading and DiaA binding. When excessive amounts of DnaA F46A were included in this crude replicative system, residual DnaA F46A activity was observed (Fig. 6F). This might be related to a slight residual activity of DnaA F46A in DnaB loading (Fig. 5, B and C) and also to the presence of factors, such as chaperons, in the crude replicative extract. Furthermore, the DnaA F46A activity was insensitive to DiaA even when DnaA F46A and DiaA were included in excess (Fig. 6, F and G). These data support the important role for DnaA Phe-46 in interactions with DiaA and DnaB during the replication reactions.
In this crude replicative system, DiaA stimulated wild-type DnaA replication activity but never inhibited it even when DiaA was present in excess (14, 15) (Fig. 6G). Limited amounts of DiaA stimulated replication, indicating that initiation complexes should interact with DiaA under these experimental conditions. These features are consistent with the in vivo function of DiaA but differ from the in vitro data obtained from systems using purified proteins for DnaB loading and minichromosomal replication as described above. We suggest that a DiaA regulatory factor is present in both the crude fraction and in vivo, which coordinates the interactions of DiaA and the DnaBC complexes with the DnaA-oriC complexes during the initiation processes (see “Discussion”).
In Vivo Initiation Regulation Requires DnaA Phe-46
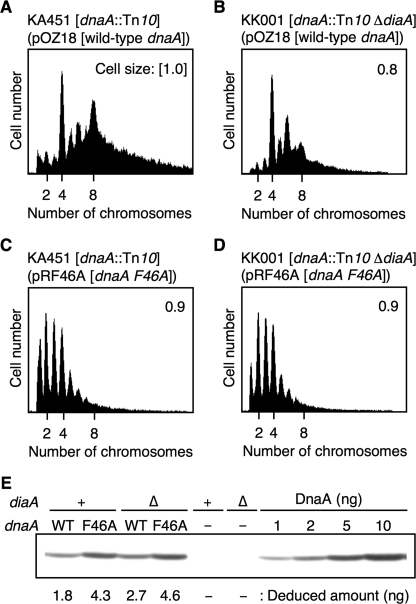
To determine the in vivo activity of DnaA F46A, we performed dnaA complementation experiments using the strain KA451 [dnaA::Tn10 rnhA::cat]. In the absence of rnhA, the chromosome is replicated using DnaA-independent alternative origins (35). The introduction of a low copy number mini-R plasmid (pOZ18), bearing the wild-type dnaA and rnhA genes, into KA451 cells leads to repression of the alternative origins and promotion of initiation at oriC on the chromosome (29). When a dnaA-defective derivative of pOZ18 (pRRNH) was introduced into KA451, the resultant transformants were unable to form colonies (Table 1). When we transformed KA451 with pOZ18 derivative pRF46A, which carries the dnaA F46A allele instead of the wild-type allele, the transformation efficiency was similar to that of pOZ18 (Table 1), although the initial rate of colony formation was slightly slower than that of KA451 transformed with pOZ18. Similar properties were observed when we transformed a KA451 derivative diaA-disrupted strain KK001 [ΔdiaA rnhA::cat dnaA::Tn10] with pRF46A (Table 1). When wild-type control (KH5402-1) cells were transformed with pRF46A, the inhibition of colony formation was not observed (Table 1). These results are consistent with our previous report (23) and with the data from the crude in vitro systems (Fig. 6, F and G). The residual activity of DnaA F46A in vivo might be enhanced by the effect of chaperon proteins and/or an increase in the cellular levels of DnaA F46A (see below).
TABLE 1.
Complementation tests of DnaA F46A in vivo
KH5402-1 (wild type) cells were transformed with mini-R derivative plasmid bearing wild-type rnhA and wild-type dnaA, dnaA F46A allele, or none and incubated at 30 °C for 20 h on LB agar plates containing thymine (50 μg/ml) and ampicillin (100 μg/ml). KA451 [dnaA ::Tn10 rnhA ::cat] and KK001 [dnaA ::Tn10 rnhA ::cat ΔdiaA] cells were similarly transformed and incubated for 36 h as above.
| Strain | Relevant genotype |
Transformation efficiency |
||||
|---|---|---|---|---|---|---|
| rnhA | dnaA | diaA | pRRNH (none) | pOZ18 (wild-type dnaA) | pRF46A(dnaA F46A) | |
| ×106 transformants/μg DNA | ||||||
| KH5402-1 | + | + | + | 1.4 | 1.1 | 1.4 |
| KA451 | − | − | + | <2.8 × 10−4 | 1.5 | 0.68 |
| KK001 | − | − | Δ | <2.8 × 10−4 | 1.9 | 0.41 |
Next, we performed flow cytometry analysis. To determine the oriC copy number in growing cells, cells were incubated in the presence of rifampicin and cephalexin for run-out replication and inhibition of cell division, followed by flow cytometry analysis. For this analysis, we first used KA451 [dnaA::Tn10] cells carrying pOZ18 [wild-type dnaA]. Cells were grown exponentially at 30 °C in LB medium. We found that most cells contained four or eight sister oriC copies per cell (Fig. 7A), although some contained five to seven oriC copies per cell. This asynchronous initiation might be caused by insufficient repression of alternative origins and/or altered transcriptional regulation of dnaA on the plasmid.
FIGURE 7.
Replication initiation activities of DnaA F46A in vivo. A–D, cells were grown exponentially at 30 °C in LB medium containing thymine (50 μg/ml) and ampicillin (50 μg/ml) and were further incubated for 4 h in the presence of rifampicin and cephalexin. Cellular DNA was then quantified using flow cytometry, and the equivalent chromosome number is indicated. The relative cell size was also measured using cells harvested at the time of drug addition, and the mean cell size is indicated as relative values when the KA451 cell size, carrying pOZ18, is defined as 1. A, KA451 [dnaA::Tn10] carrying pOZ18 (wild-type dnaA). B, KA451 carrying pRF46A [dnaA F46A]. C, KK001 [dnaA::Tn10 ΔdiaA] carrying pOZ18. D, KK001 carrying pRF46A. E, cellular DnaA amounts were determined by immunoblot analysis. KA451 (+) and KK001 (Δ) cells carrying pOZ18 (WT), pRF46A (F46A), or pMZ1 (vector; −) were grown under the same conditions as those used for preparation of the flow cytometry samples. A portion of each culture was used for SDS-PAGE, followed by immunoblot analysis. KA451 and KK001 cells, carrying pMZ1, were used as a background control. The indicated amounts of purified DnaA protein were mixed with whole cell extract from KA451 cells carrying pMZ1 for a quantitative standard.
KK001 [ΔdiaA dnaA::Tn10] cells carrying pOZ18 grew at a similar rate (∼95 min/generation) to KA451 cells carrying pOZ18, and predominantly contained four and six oriC copies per cell (Fig. 7B). There was a severe reduction in the numbers of KK001 cells containing eight oriC copies compared with those of KA451 cells carrying pOZ18. The mean cell size of KK001 cells carrying pOZ18 was only slightly smaller than that of KA451 carrying pOZ18. The amount of DnaA protein present in KK001 cells carrying pOZ18 was not reduced compared with that in KA451 cells bearing pOZ18 (Fig. 7E). These results indicate that DiaA stimulates the initiation of chromosomal replication in this strain system.
Based on the above results, we analyzed KA451 and KK001 cells both carrying pRF46A [dnaA F46A]. These cells grew in a log phase at a similar rate to KA451 cells carrying pOZ18. In both cases, the majority of cells contained only one to four oriC copies per cell (Fig. 7, C and D). The mean cell size of both KA451 and KK001 cells carrying pRF46A was similar to that for KA451 cells carrying pOZ18 (Fig. 7, C and D). The amounts of DnaA F46A protein in these cells did not decrease, rather the cellular DnaA content in these strains increased ∼2-fold compared with that in KA451 cells carrying pOZ18 (Fig. 7E). These results indicate that DnaA F46A activity during initiation of replication in vivo is impaired, consistent with the current in vitro results indicating defects in the DiaA interaction and DnaB loading. The in vivo interaction of DnaA F46A with DnaB would occur at an impaired efficiency as seen in the in vitro crude extract experiments (Fig. 6F). The slight increase in the cellular amounts of DnaA F46A could be caused by a defect in the transcriptional autoregulation of the dnaA gene (36). Moreover, the peak pattern of KK001 cells carrying pRF46A was similar to that for KA451 cells carrying pRF46A (Fig. 7, C and D). This is consistent with a defect in the DiaA-DnaA F46A interaction.
DISCUSSION
In this study, we used NMR analysis and a pulldown assay to demonstrate that the presence of DnaA Phe-46, which occurs within DnaA domain I, is a prerequisite for DiaA binding (Figs. 1 and 2). The DnaA Phe-46-DiaA interaction is required to stimulate the formation of ATP-DnaA-specific initiation complexes and open complexes (Fig. 3C and Fig. 4B), consistent with the in vitro replication activity and in vivo analysis (Fig. 6, F and G, and Fig. 7). A structural model for the DiaA-DnaA domain I complex provides a practical structural basis for understanding the mechanism involved in the DnaA assembly on oriC (Fig. 1E). Moreover, we found that DnaA Phe-46 is also important for DnaB helicase loading onto open complexes and for direct binding of DnaB or the DnaBC complex to DnaA-oriC complexes (Fig. 5, B and C, and Fig. 6, A and B), which is also consistent with our in vitro and in vivo data (Fig. 6, F and G, and Fig. 7). We therefore propose that DnaA Phe-46 plays two key roles. First, it is involved in the DiaA-dependent regulation of the initial complex formation; and second, it is involved in the loading of DnaB onto the complexes for the timely initiation of chromosomal replication.
The residue corresponding to E. coli DnaA Phe-46 is extremely highly conserved in eubacterial DnaA orthologs (23). E. coli DiaA orthologs are also widely conserved among eubacterial species (15). Notably, the Helicobacter pylori DiaA ortholog HobA is reported to be essential in cell growth and is known to directly bind the cognate DnaA and to stimulate the assembly of the DnaA molecules on the cognate oriC (37–39). Similarly, it would be possible that dynamics of the initial complexes during replicative helicase loading as well as DiaA-controlled DnaA assembly are conserved among eubacterial species.
We further suggest that DiaA and DnaB interact with only a subgroup of DnaA molecules assembled on oriC (Figs. 3–5). The compositions of DnaA-binding sites within oriC are highly elaborately organized, which would lead to heterogeneous substructures within an initiation complex. We speculate that only a specific DnaA subgroup in the complex might expose domain I to a position available for the protein loading. Consistently, we found that DiaA inhibits the DnaA-DnaB interaction and DnaB loading onto DnaA multimers on oriC (Fig. 6). These observations support the simple idea that DiaA and DnaB interact with a common subgroup of DnaA molecules assembled at the origin. In a footprint analysis, DiaA promoted the cooperative binding of ATP-DnaA, and this was especially effective on the τ1-to-I2 ADLAS within oriC (Fig. 3). This region is located near the DUE, which would be favorable for DnaB loading onto ssDUE. DiaA and DnaB might commonly use a subgroup of DnaA molecules located at this region.
The timing of DnaB loading has been thought to be regulated solely by open complex formation. This study, however, indicated that DnaBC stably complexes with DnaA multimers in the absence of DiaA, and this does not always require open complexes (Fig. 6, A and B). We suggest a novel possibility that DiaA plays a negative regulatory role in DnaB helicase loading, following its positive role in DnaA assembly in the initiation process. DiaA-controlled DnaB loading inhibition may be related to our previous in vivo data, which showed that a moderate overproduction of DiaA using pBR322-diaA plasmid retards the onset of initiation of chromosomal replication, thereby resulting in asynchronous initiations (14). We can speculate that this negative role is important in ensuring the stepwise progression of initiation reactions and the timely initiation of chromosomal replication. As such, it is conceivable that the occurrence of timely initiation, including synchronous initiations of multiple origins, is regulated strictly using multiple regulatory points, including the increase of cellular ATP-DnaA levels, the assembly of ATP-DnaA molecules on oriC, and DnaB helicase loading.
It is likely that a DiaA-regulating factor is present in vivo, because we observed that a crude extract eliminated the DiaA inhibition of DnaB loading onto open complexes, and it is also known that minichromosomes can efficiently replicate in a crude extract, including DiaA (Fig. 6G) (14, 15). In analyses on subcellular localization, it has been suggested that DiaA co-localizes with oriC during a specific moment only in the cell cycle.3 A DiaA-regulating factor might release DiaA from DnaA-oriC complexes in a timely manner after DiaA-dependent DnaA assembly, which might determine the precise timing of DnaB helicase loading onto oriC during the cell cycle. Alternatively, a DiaA-regulating factor might promote overall structural change of DnaA-oriC complexes, thereby allowing another DnaA subgroup to become available to DnaB loading.
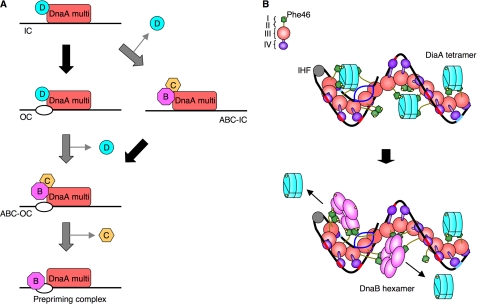
Based on the analyses mentioned above, we proposed novel models to explain the mechanisms controlling the initiation of replication and its regulation (Fig. 8A). When the cellular ATP-DnaA level is elevated, DiaA promotes the formation of a stable initiation complex by the assembly of ATP-DnaA molecules on oriC, through its interaction with DnaA domain I, thus resulting in the formation of an open complex. At this stage, a DiaA-regulatory factor releases DiaA from the complex in a regulated and timely manner. Subsequently, DnaBC complexes are loaded onto the open complex (ABC-open complex), followed by prepriming complex formation (Fig. 8A, left). Alternatively, DiaA may be released immediately after the functional initiation complexes are formed, which allows the association of DnaBC complexes with the initiation complex (ABC-initiation complex), followed by open complex formation and DnaB loading onto the unwound DNA strands (Fig. 8A, right). As mentioned above, a DiaA-regulating factor may change overall structure of the initial complex, allowing DnaB loading.
FIGURE 8.
Models for the DnaA initial complexes, including DiaA or DnaB. A, model for the basic mechanisms involved in the initiation of DNA replication. A DnaA multimer (DnaA multi) complexed with DiaA (D) is formed on oriC, followed by the timely release of DiaA and the loading of complexes of DnaB (B) and DnaC (C) (see text for details). The protein factors are depicted in different colors. IC, initiation complex; OC, open complex; ABC-IC, initiation complex including DnaABC; ABC-OC, open complex including DnaABC. The black arrows indicate formation of an open complex. B, model for the higher order structures of the initial complexes, including DiaA or DnaB. DiaA stimulates DnaA assembly on oriC, resulting in a spiral of DnaA multimers and the interaction of DiaA with specific DnaA subgroups (top). The identical DnaA subgroups are used also by the DnaB hexamer helicase following dissociation of DiaA (bottom) (see text for details). DnaA domains I–IV, DnaA Phe-46, DiaA tetramer, DnaA binding patch of DiaA, integration host factor (IHF), and DnaB hexamer are indicated in different colors. DUE (blue), DnaA boxes (R1 and R4 sites) (aqua), and ADLAS (DnaA boxes R2/R3/M, τ and I sites) (red) within oriC are also indicated.
We have combined all the results discussed above in a novel model for initiation complex structures (Fig. 8B). ATP-DnaA forms multimers on strong DnaA boxes and ADLAS within oriC, possibly forming a spiral shape (21, 22). DiaA tetramers intercalate into the DnaA spiral and effectively interact with subgroups of DnaA molecules being loaded onto ADLAS via DnaA domain I Phe-46, thus stimulating cooperative binding and stabilizing the DnaA multimer (Fig. 8B, top). Two or three DiaA tetramers would be included in a single initiation complex containing ∼20 DnaA molecules (15) (Fig. 2D and supplemental Fig. S1B). Other subgroups of DnaA molecules do not bind to DiaA and thus allow DnaA domain I to form self-oligomers, thereby assisting stable inter-DnaA interaction (23, 40). Simultaneously, formation of the ATP-DnaA-specific oriC complex is promoted by specific interactions between DnaA domain III mediated by the AAA+ arginine finger (Arg-285) and ATP (20). The resultant ATP-DnaA multimer is a functional initiation complex competent in DUE unwinding (Fig. 8B, top).
Following DUE unwinding, the T-rich ssDUE strand would interact with specific DnaA AAA+ domain residues (B/H motifs) on the pore surface of the DnaA spiral (21). The integration host factor could then promote interactions among ATP-DnaA molecules by bending the DNA at the binding site between the DnaA box R1 and τ1 (41, 42). After the active release of DiaA, DnaB in the DnaBC complexes would, in turn, bind effectively to an open complex using domain I Phe-46 in a DnaA subgroup that had been used for DiaA binding (Fig. 8B, bottom). Alternatively, another DnaA subgroup might become available to DnaB loading by overall structural change of the complex as mentioned above. Also, unwinding of the DUE might follow DnaBC loading as explained above.
On eukaryotic replication origins, MCM helicase binds directly to the origin recognition complex-Cdc6 complex in a Cdt1-mediated manner, forming the pre-replicative complex (43). Multiple proteins such as Cdc45, Sld3, and Dpb11 are loaded onto the pre-replicative complex and are mediated by GINS (44). Notably, GINS is a tetramer complex consisting of subunits with mutually similar tertiary structures (45–47). Moreover, the structural modes involved in complex formation by these subunits are similar between DiaA and GINS. These similarities between DiaA and GINS could provide an insight into the evolutionary conservation and divergence of initiation mechanisms.
Supplementary Material
This work was supported by grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Technology and Science of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and additional references.
S. Shinozaki, T. Hatano, H. Niki, and T. Katayama, unpublished data.
- DUE
- DNA unwinding element
- AAA
- ATPases associated with a variety of cellular activities
- ss
- single-stranded
- ADLAS
- ATP-DnaA-preferential low affinity sites
- HSQC
- heteronuclear single quantum correlation
- RIDA
- regulatory inactivation of DnaA
- SSB
- single-strand binding protein.
REFERENCES
- 1.Messer W. (2002) FEMS Microbiol. Rev. 26, 355–374 [DOI] [PubMed] [Google Scholar]
- 2.Mott M. L., Berger J. M. (2007) Nat. Rev. Microbiol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 3.Zakrzewska-Czerwiñska J., Jakimowicz D., Zawilak-Pawlik A., Messer W. (2007) FEMS Microbiol. Rev. 31, 378–387 [DOI] [PubMed] [Google Scholar]
- 4.Sekimizu K., Bramhill D., Kornberg A. (1987) Cell 50, 259–265 [DOI] [PubMed] [Google Scholar]
- 5.Bramhill D., Kornberg A. (1988) Cell 52, 743–755 [DOI] [PubMed] [Google Scholar]
- 6.Marszalek J., Kaguni J. M. (1994) J. Biol. Chem. 269, 4883–4890 [PubMed] [Google Scholar]
- 7.Fang L., Davey M. J., O'Donnell M. (1999) Mol. Cell 4, 541–553 [DOI] [PubMed] [Google Scholar]
- 8.O'Donnell M. (2006) J. Biol. Chem. 281, 10653–10656 [DOI] [PubMed] [Google Scholar]
- 9.Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T. (1999) EMBO J. 18, 6642–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama T., Kubota T., Kurokawa K., Crooke E., Sekimizu K. (1998) Cell 94, 61–71 [DOI] [PubMed] [Google Scholar]
- 11.Kato J., Katayama T. (2001) EMBO J. 20, 4253–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su'etsugu M., Shimuta T. R., Ishida T., Kawakami H., Katayama T. (2005) J. Biol. Chem. 280, 6528–6536 [DOI] [PubMed] [Google Scholar]
- 13.Su'etsugu M., Nakamura K., Keyamura K., Kudo Y., Katayama T. (2008) J. Biol. Chem. 283, 36118–36131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishida T., Akimitsu N., Kashioka T., Hatano M., Kubota T., Ogata Y., Sekimizu K., Katayama T. (2004) J. Biol. Chem. 279, 45546–45555 [DOI] [PubMed] [Google Scholar]
- 15.Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su'etsugu M., Fujimitsu K., Kagawa W., Yokoyama S., Kurumizaka H., Katayama T. (2007) Genes Dev. 21, 2083–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S. (2003) Nucleic Acids Res. 31, 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neuwald A. F., Aravind L., Spouge J. L., Koonin E. V. (1999) Genome Res. 9, 27–43 [PubMed] [Google Scholar]
- 18.Iyer L. M., Leipe D. D., Koonin E. V., Aravind L. (2004) J. Struct. Biol. 146, 11–31 [DOI] [PubMed] [Google Scholar]
- 19.Felczak M. M., Kaguni J. M. (2004) J. Biol. Chem. 279, 51156–51162 [DOI] [PubMed] [Google Scholar]
- 20.Kawakami H., Keyamura K., Katayama T. (2005) J. Biol. Chem. 280, 27420–27430 [DOI] [PubMed] [Google Scholar]
- 21.Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S. Y., Yokoyama S., Kurumizaka H., Katayama T. (2008) J. Biol. Chem. 283, 8351–8362 [DOI] [PubMed] [Google Scholar]
- 22.Erzberger J. P., Mott M. L., Berger J. M. (2006) Nat. Struct. Mol. Biol. 13, 676–683 [DOI] [PubMed] [Google Scholar]
- 23.Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T. (2007) J. Biol. Chem. 282, 17816–17827 [DOI] [PubMed] [Google Scholar]
- 24.Nozaki S., Ogawa T. (2008) Microbiology 154, 3379–3384 [DOI] [PubMed] [Google Scholar]
- 25.Sutton M. D., Carr K. M., Vicente M., Kaguni J. M. (1998) J. Biol. Chem. 273, 34255–34262 [DOI] [PubMed] [Google Scholar]
- 26.Seitz H., Weigel C., Messer W. (2000) Mol. Microbiol. 37, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 27.McGarry K. C., Ryan V. T., Grimwade J. E., Leonard A. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2811–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abe Y., Watanabe N., Yoshida Y., Ebata F., Katayama T., Ueda T. (2007) Biomol. NMR Assign. 1, 57–59 [DOI] [PubMed] [Google Scholar]
- 29.Kawakami H., Ozaki S., Suzuki S., Nakamura K., Senriuchi T., Su'etsugu M., Fujimitsu K., Katayama T. (2006) Mol. Microbiol. 62, 1310–1324 [DOI] [PubMed] [Google Scholar]
- 30.Fujimitsu K., Su'etsugu M., Yamaguchi Y., Mazda K., Fu N., Kawakami H., Katayama T. (2008) J. Bacteriol. 190, 5368–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang P., Marians K. J. (2000) J. Biol. Chem. 275, 26187–26195 [DOI] [PubMed] [Google Scholar]
- 32.Riber L., Fujimitsu K., Katayama T., Løbner-Olesen A. (2009) Mol. Microbiol. 71, 107–122 [DOI] [PubMed] [Google Scholar]
- 33.Masai H., Nomura N., Arai K. (1990) J. Biol. Chem. 265, 15134–15144 [PubMed] [Google Scholar]
- 34.Baker T. A., Sekimizu K., Funnell B. E., Kornberg A. (1986) Cell 45, 53–64 [DOI] [PubMed] [Google Scholar]
- 35.Kogoma T. (1997) Microbiol. Mol. Biol. Rev. 61, 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun R. E., O'Day K., Wright A. (1985) Cell 40, 159–169 [DOI] [PubMed] [Google Scholar]
- 37.Terradot L., Durnell N., Li M., Li M., Ory J., Labigne A., Legrain P., Colland F., Waksman G. (2004) Mol. Cell. Proteomics 3, 809–819 [DOI] [PubMed] [Google Scholar]
- 38.Natrajan G., Hall D. R., Thompson A. C., Gutsche I., Terradot L. (2007) Mol. Microbiol. 65, 995–1005 [DOI] [PubMed] [Google Scholar]
- 39.Zawilak-Pawlik A., Kois A., Stingl K., Boneca I. G., Skrobuk P., Piotr J., Lurz R., Zakrzewska-Czerwiñska J., Labigne A. (2007) Mol. Microbiol. 65, 979–994 [DOI] [PubMed] [Google Scholar]
- 40.Felczak M. M., Simmons L. A., Kaguni J. M. (2005) J. Biol. Chem. 280, 24627–24633 [DOI] [PubMed] [Google Scholar]
- 41.Polaczek P., Kwan K., Liberies D. A., Campbell J. L. (1997) Mol. Microbiol. 26, 261–275 [DOI] [PubMed] [Google Scholar]
- 42.Ryan V. T., Grimwade J. E., Nievera C. J., Leonard A. C. (2002) Mol. Microbiol. 46, 113–124 [DOI] [PubMed] [Google Scholar]
- 43.Bell S. P., Dutta A. (2002) Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 44.Labib K., Gambus A. (2007) Trends Cell Biol. 17, 271–278 [DOI] [PubMed] [Google Scholar]
- 45.Chang Y. P., Wang G., Bermudez V., Hurwitz J., Chen X. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12685–12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi J. M., Lim H. S., Kim J. J., Song O. K., Cho Y. (2007) Genes Dev. 21, 1316–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamada K., Kubota Y., Arata T., Shindo Y., Hanaoka F. (2007) Nat. Struct. Mol. Biol. 14, 388–396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.