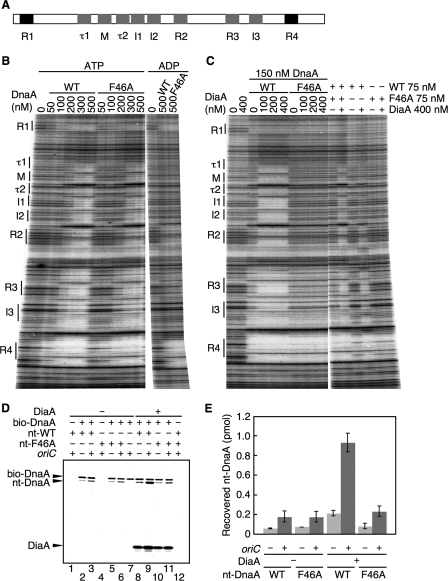
FIGURE 3.
DnaA F46A is defective in DiaA-dependent formation of DnaA-oriC complexes. A, structure of oriC DnaA-binding region. The positions of the DnaA-binding sequences are indicated. High affinity sites (R1 and R4) and ADLAS (R2, R3, M, and I/τ sites) are colored in black and gray, respectively. B, amounts of wild-type DnaA (WT) and DnaA F46A (F46A) indicated were incubated at 30 °C for 10 min in buffer containing a 32P-end-labeled oriC fragment and 3 mm ATP or ADP, followed by DNase I footprint analysis using 5% sequencing gel electrophoresis. The positions of the DnaA-binding sequences (boxes R1–R4 and M; sites I1–3 and τ1–2) are indicated. C, indicated amounts of DiaA (as monomers) were incubated at 30 °C for 10 min, in the presence of 3 mm ATP, with the indicated amounts of wild-type DnaA and/or DnaA F46A, followed by DNase I footprint analysis as described for B. D, reactions were incubated on ice for 15 min in the presence (+) or absence (−) of His-DiaA (500 nm; 5 pmol as monomer), bio-DnaA (250 nm; 2.5 pmol), nontagged wild-type DnaA (nt-WT) (250 nm; 2.5 pmol), DnaA F46A (nt-F46A) (250 nm; 2.5 pmol), oriC fragment (10 nm; 100 fmol). Proteins bound to streptavidin beads were isolated and analyzed by SDS-10% PAGE and silver staining. Nt-DnaA, nontagged DnaA. E, recovered amounts of nt-WT and nt-F46A were determined using standard curves, and the average values from two independent experiments were indicated.