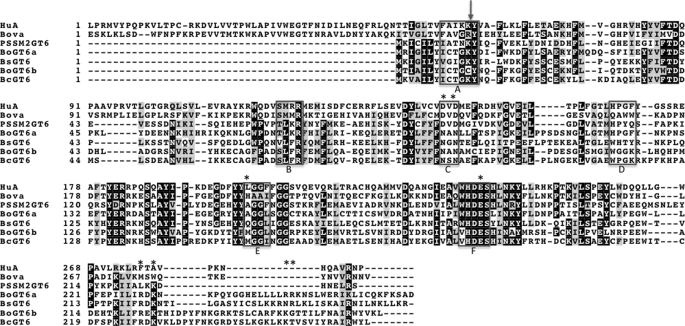
FIGURE 1.
An alignment of selected bacterial, cyanophage and mammalian GT6 amino acid sequences. Abbreviations and Interpro sequence IDs (in parentheses) are as follows. HuA, human histo-blood group A synthase (A1EAJ6); Bova, bovine α1,3-galactosyltransferase (P14769); PSSM2, cyanophage PSSM-2 (Q58M87); Bs, B. stercoris (B0NSM3); Bo1, B. ovatus GT1 (A7LVT2); Bo2, B. ovatus GT2 (A7M0P3); Bc, B. caccae (A5ZC71). The boxed regions in the alignment identify regions that have been shown to be involved in interactions with substrates and cofactor and in catalysis in bovine α1,3-galactosyltransferase and histo-blood group A and B enzymes. These are labeled (below) as follows. A, interactions with uracil; B, interactions with the galactose moiety of UDP-Gal; C, interactions with Mn2+, phosphates, and galactose; D, interactions with acceptor substrate; E, interactions with Gal or GalNAc of donor substrate; F, interactions with monosaccharide of donor substrate and acceptor and catalysis; The arrow (above) denotes the intron/exon boundary in vertebrate GT6s, and the asterisks indicate the residues in BoGT6a that were subjected to mutagenesis.