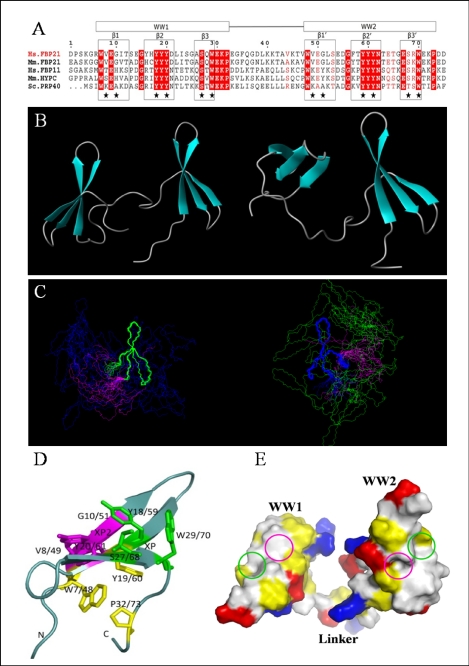
FIGURE 5.
Solution structure of the tandem WW domains of FBP21. A, sequence alignment of FBP21 tandem WW domains with its orthologues as follows: Hs.FBP21 and Hs.FBP11 from Homo sapiens; Mm.FBP21 and Mm.HYPC from Mus musculus; and Sc.PRP40 from Saccharomyces cerevisiae. The WW domain boundaries are indicated by boxes at the top, and the secondary structure elements are boxed and indicated above the alignment. Residues identical in all five sequences are shown in red columns, and conserved residues are shown in red letters. Stars indicate residues forming binding interface. The sequence alignment was made using ClustalW, and the panel was generated by ESPript 2.2. B, ribbon representation of two structures in different interdomain orientations with secondary elements of WW domains shown as turquoise arrows. C, stereoview of the 20 lowest energy NMR structures of FBP21 tandem WW domains, superimposed on backbone atoms of WW1 (blue) and WW2 (green), respectively, and the flexible linker is shown in magenta. The structures cannot be superimposed over the entire molecule due to the flexible interdomain motion. D, view of the binding surface on the tandem WW domains of FBP21. Residues that form the XP and XP2 grooves are shown as green and purple sticks, respectively. The side chains of residues forming a hydrophobic core that stabilizes the fold are shown in yellow. The figures were made using MOLMOL and PyMol. E, space-filling structure of the WW domains of FBP21. The molecular surface is colored according to electric and hydrophobic potential. Blue colors represent positive charges; red colors are negative charges; neutral areas are gray, and yellow represents hydrophobic residues. The green and purple circles represent the XP and XP2 grooves, respectively.