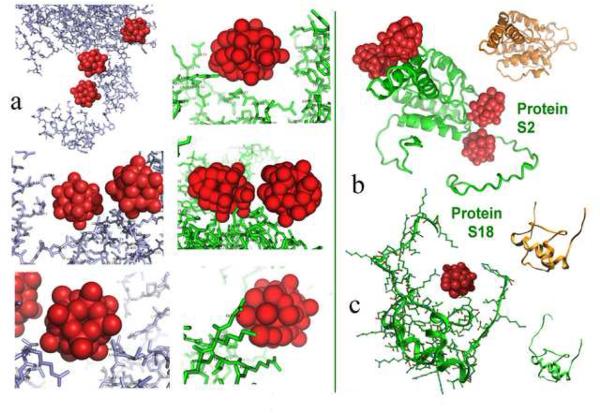
Figure 2.
(a) Typical heteropolytungstate interactions in both T30S and the LDL receptor extracellular domain. Note the similarity of the cluster “nests” created in the two systems. Also common is the high density of the tungsten clusters, which frequently appear very close to each other. (b) examples for conformational alterations induced by the cluster binding in two r-proteins: S2 and S18. For comparison the structures of the same proteins in the nontungstenated T30S [3] are also shown (in gold).
In all the heteropolytungstate clusters are shown as groups of 12 [83] or 18 [2] red balls, according to the number of the W atoms in the respective cluster 9 (Na3PW12O40 and (NH4)6(P2W18O62)14H2O). The LDL receptor extracellular domain is shown in metal blue. R-proteins S2 and S18 are shown in green in the tungstenated T30S [2] and in gold in the nontungstenated T30S [3]. Amino acid numbering for proteins of T30S is according to E. coli. In items showing highly dense regions, the numbering was removed, for clarity.
(b and c) show the structures of proteins S2 (b) and S18 (c) within the tungstenated [2] and non-tungstenated [3] T30S crystals in. For both the main chain is shown (in green and gold respectively). Note the marked difference in the conformation of the termini of protein S2. Also, note that all atoms of protein S18 are resolved in the tungstenated crystal, including those embracing the cluster, whereas over a dozen aminoacids are disordered in the nontungstenated crystal. The all-atom presentation is also shown, for highlighting typical “nest” architecture.