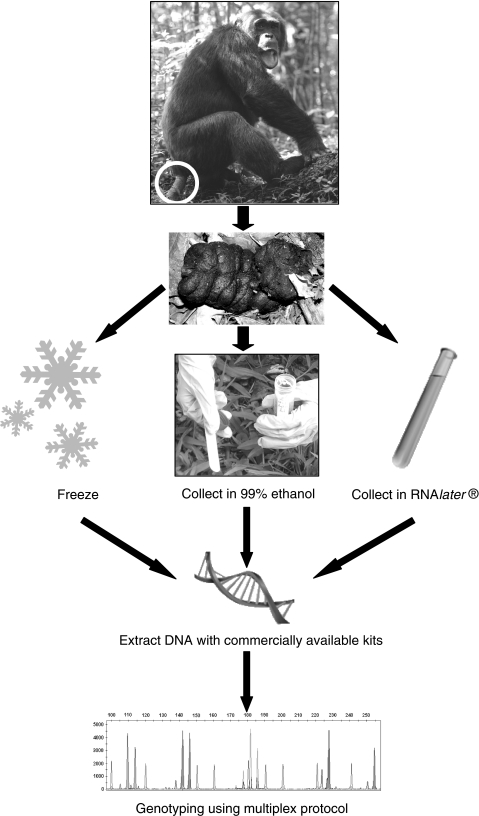
Fig. 1.
Flow chart of sample processing from collection to genotyping. For sample collection it is best to target fresh dung (less than 24 h old). For known individuals we recommend collection of two or three samples to confirm identity. Three methods of sample collection provide good results. If conditions allow, one can freeze the sample at −20°C or in liquid nitrogen. Because freezing is usually difficult in the field, collection in RNAlater or using the two-step collection method (Nsubuga et al. 2004) is recommended. Researchers interested in the study of pathogens may prefer to collect samples in RNAlater (Leendertz et al. 2006). Such samples, of course, also contain DNA of the study individual and can be used for population genetic studies (Eriksson et al. 2004). The two-step collection method has been successfully used for a variety of species (great apes, macaques, black-and-white colobus). We recommend collection of approximately 5 g (equivalent to a teaspoonful) of fresh dung and immersion in ~35 ml 90–99% ethanol. The ratio of dung to ethanol should be approximately 1:7; lower ratios can result in lower genotyping success. After storage in ethanol for 24 h, the sample is transferred on to silica beads for complete desiccation. Whatever method is used for sample collection, DNA is extracted from dung samples either using commercially available kits (Bradley et al. 2000) or by use of other extraction methods (Vallet et al. 2008). Multiplexing procedures can increase the speed and accuracy of genotyping and help to save the valuable genetic material by utilizing only small amounts of template (Lampa et al. 2008; Arandjelovic et al. 2008). Pictures are courtesy of K. Langergraber, R. Ikfuingei, T. Harris, and M. Arandjelovic