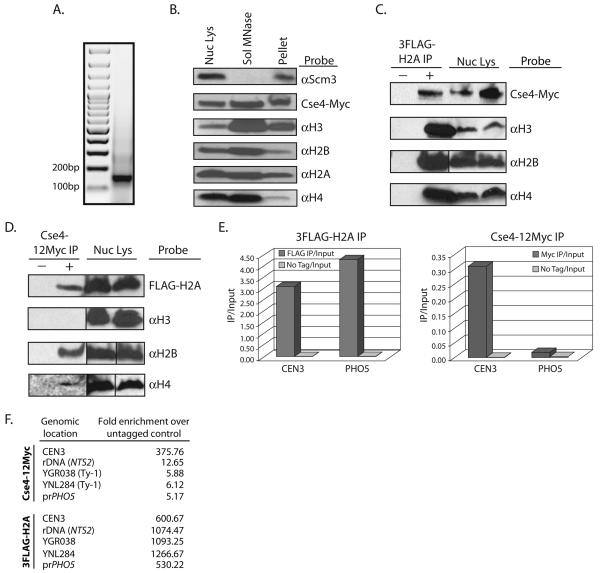
Figure 4.
Co-Immunoprecipitation of histones from MNase-solubilized chromatin. Nuclear lysates were made from a strain expressing both 3FLAG-H2A and Cse4-12Myc (RC188). The chromatin fraction was pelleted and treated with MNase. Western blots were probed with each antibody listed. For each antibody, the lanes are from a single exposure of the same gel. Vertical lines indicate that intervening lanes were cropped. A. DNA was isolated from the MNase solubilized chromatin and was visualized with ethidium bromide on an agarose gel to ensure a majority of mononucleosomes. B. Total nuclear lysate, the MNase-solubilized chromatin fraction, and the insoluble chromatin pellet after MNase treatment were loaded for Western blot analysis. C. Co-immunoprecipitation was performed using the MNase-solubilized chromatin from strain RC188. αFLAG-conjugated beads were used to pulldown 3FLAG-H2A, and the pulldown was probed using the antibodies listed. The negative control pulldown was performed using chromatin from a strain lacking 3FLAG-H2A (SBY617). D. αMyc-conjugated beads were used to pulldown Cse4-12Myc from chromatin made from RC188, and the pulldown was probed using the antibodies listed. The negative control pulldown was performed using chromatin from a strain lacking Cse4-12Myc (RC177). E. DNA was isolated from both the αFLAG and αMyc pulldowns (NChIP). qPCR was then performed to look for enrichment of DNA at either CEN3 or at the site of a well-positioned canonical nucleosome at the PHO5 promoter. Cse4 is significantly enriched at CEN3 when compared to the PHO5 promoter. Levels of 3FLAG-H2A are similar at both sites. F. qPCR signal from immunoprecipitated Cse4-12Myc and 3FLAG-H2A compared to an untagged control strain at CEN3, the rDNA, Ty elements and the PHO5 promoter. Fold enrichment was calculated by dividing IP/input ratios from the untagged control by the IP/input ratios of the epitope-tagged samples.