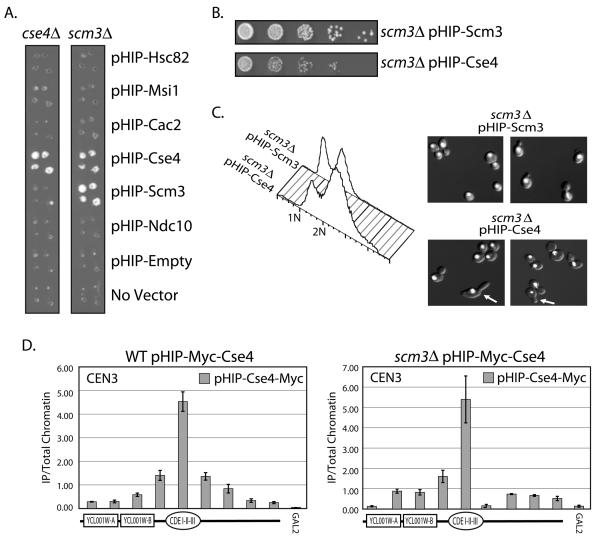
Figure 5.
Suppression of the scm3Δ strain. The SCM3/scm3Δ (RC204) or CSE4/cse4Δ (RC203) heterozygous diploid knockout “Magic Marker” strain was transformed with plasmids from the HIP overexpression library. A. Sporulated cultures were pinned in quadruplicate onto medium which allowed the recovery of haploid yeast containing either scm3Δ or cse4Δ and the plasmid indicated. B. Dilution assay comparing the growth of scm3Δ covered by either pHIP-Scm3 or pHIP-Cse4 on Gal-Ura medium. C. FACscan analysis and DAPI staining of the scm3Δ strain covered by pHIP-Scm3 and scm3Δ covered by pHIP-Cse4. DAPI stained DNA is shown in white. Arrows indicate cells with abnormal morphology (elongated buds). D. qPCR analysis of pHIP-Myc-Cse4 localization was performed in both a wild type (WT) Scm3 strain containing the pHIP-Myc-Cse4 plasmid (RC205) and a scm3Δ strain containing the pHIP-Myc-Cse4 plasmid (RC192). The primers used amplify a +/− 2kb region around CEN3. A depiction of the features within the region is shown below each histogram. GAL2 serves as a negative control for Cse4 localization. Error bars represent +/− the average deviation of biological replicates.