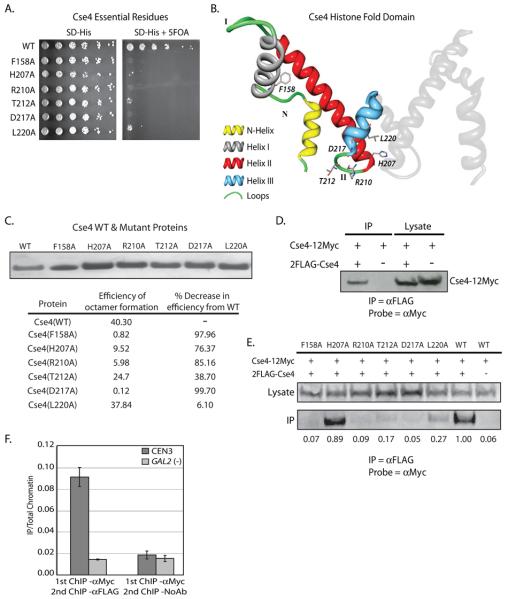
Figure 7.
Dimerization of Cse4. Cse4 was cloned into pRS413 and subjected to site directed mutagenesis which mutated each individual amino acid to an alanine. Each mutated plasmid in the collection was transformed into a haploid cse4Δ strain containing another plasmid with a wild type copy of Cse4, and a plasmid-shuffle assay was performed. A. Growth on 5-FOA identified six single alanine substitutions that do not support growth in the cse4Δ background. B. Using the modeled Cse4 crystal structure as a guide (Bloom et al., 2006), the location of each of the 6 lethal point mutants was mapped. One molecule of Cse4 histone fold domain from the predicted Cse4 octamer crystal structure is shown in color and the essential residues are indicated. The second molecule of Cse4 is shown in grey. Five of six of the lethal point mutations lie in close proximity in either Loop II or Helix III. C. Each of the Cse4 mutants identified in our alanine scanning mutagenesis (7A) was purified from E. coli inclusion bodies. We then used these recombinant Cse4 point mutants to reconstitute octamers in vitro. Octamers were subjected to gel filtration chromatography. The efficiency of octamer formation was calculated by dividing the amount (mg) of octamer recovered by the amount of input histones (Cse4, H2A, H2B, H4). D. Co-immunoprecipitation was performed using whole cell extracts (WCE) isolated from a strain which expresses both Cse4-12Myc and 2FLAG-Cse4 (MM118). αFLAG-conjugated beads were used to pulldown 2FLAG-Cse4 from WCE, and the pulldown was probed by Western blotting with the αMyc antibody. The negative control pulldown was performed using WCE from a strain lacking the 2FLAG tag on Cse4 (MM117). E. Co-immunoprecipitation was performed from WCE isolated from a strain which expresses both Cse4-12Myc and a 2FLAG-Cse4 point mutant identified in the alanine scanning mutagenesis (MM111-116), WT 2FLAG-Cse4 (MM118), or Cse4 without a FLAG tag (MM117). αFLAG-conjugated beads were used to pulldown 2FLAG-Cse4 from WCE, and the pulldown was probed by Western blotting with the αMyc antibody. The values below each lane of the blot represent quantification of the IP band over the lysate band, with the WT sample set to 1. E. SeqXChIP was performed using sheared chromatin isolated from MM118. αMyc antibody was used for the 1st round of XChIP, followed by XChIP using either the αFLAG antibody or no antibody. The signal from each XChIP has been divided by the signal obtained with total chromatin. The centromeric primer pair spans ~ 350bp across CEN3. The GAL2 gene serves as a negative control for Cse4 localization. Error bars represent +/− the average deviation of biological replicates.