Fig. 6.
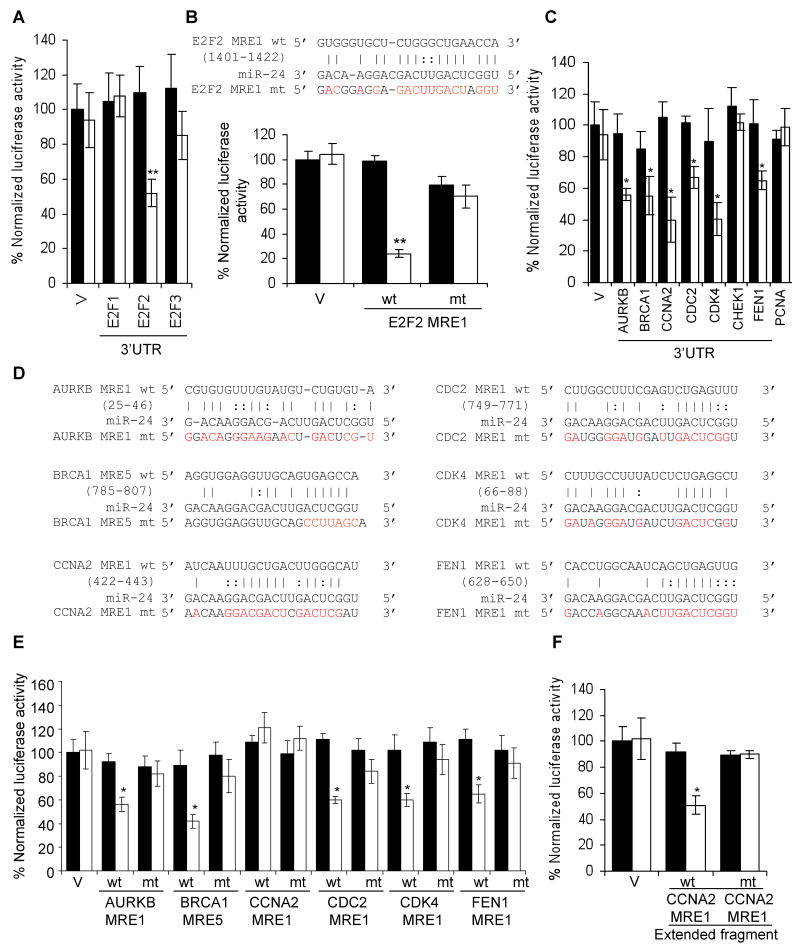
E2F2 and multiple E2F-target genes are direct targets of miR-24, recognized by “seedless” MREs. (A) miR-24 silences the expression of luciferase genes engineered with the 3′UTR of E2F2, but not with E2F1 or E2F3 3′UTRs, suggesting that E2F2 is a direct miR-24 target but E2F1 and E2F3 are down-regulated indirectly. Luciferase assays were performed in HepG2 cells over-expressing miR-24 (white) or control mimics (black). (B) miR-24 down-regulates luciferase activity of a reporter gene containing wild-type (wt) E2F2 MRE1. Mutations in the miR-24 pairing residues (mt) rescue luciferase expression (sequences and luciferase assays for candidate E2F2 wt 3′UTR MREs are shown in Suppl. Fig. 3). (C) miR-24 targets the 3′UTR of E2F-regulated genes (AURKB, BRCA1, CCNA2, CDC2, FEN1) and CDK4, a MYC-regulated gene. CHEK1 and PCNA 3′UTRs are not regulated by miR-24. HepG2 cells were co-transfected with a luciferase reporter containing the 3′UTR of the indicated gene and control miRNA (black) or synthetic miR-24 (white) for 48 hr. Expression of the unmodified luciferase vector (V) is unchanged by miR-24. (D) Predicted binding sites in the 3′UTR of genes whose 3′UTR was repressed by miR-24 in (C) and binding site mutations tested (indicated in red). (E) Expression of reporter genes containing wild-type (wt) AURKB MRE1, BRCA1 MRE5, CDC2 MRE1, CDK4 MRE1 and FEN1 MRE1 is significantly reduced upon co-transfection of HepG2 cells with miR-24 mimics (white) and not the control mimic (black). Mutations in the miR-24 pairing residues (mt) rescue luciferase expression (sequences and luciferase assays for all tested wt MREs for these genes in Suppl. Fig. 6,7). The CCNA2 MRE1 is not regulated. (F) However, miR-24 regulates a 181 nt region containing the CCNA2 MRE1 in the luciferase vector. Mutations in the binding residues of CCNA2 MRE1 within the extended sequence restore luciferase activity. Error bars in A, C and E represent mean ± SD from 3 independent experiments. **, p<0.01; *, p<0.05.