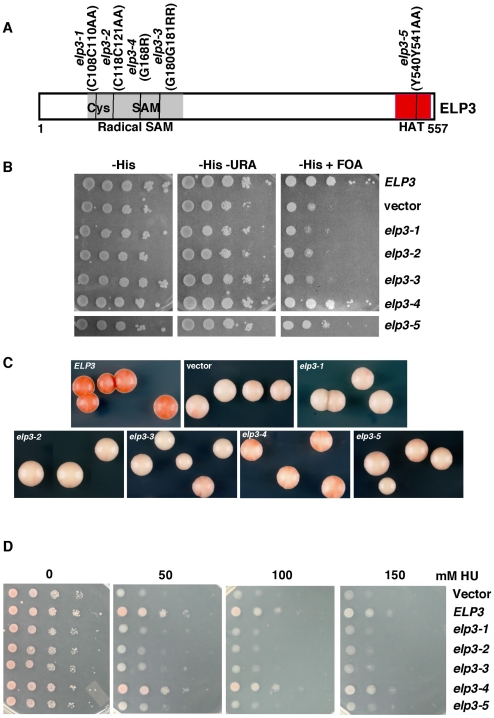
Figure 2. Cells expressing elp3 site-specific mutants with mutations at the HAT domain and the conserved Radical SAM domain of Elp3 reduce transcriptional silencing and are sensitive to HU.
(A) A schematic representation of mutations on Elp3. Highlighted are the domains that bind S-adenosylmethionine (SAM) and the signature of histone acetyltransferase (HAT). Several amino acids located in the Radical SAM domains were mutated and these included elp3-1 with cysteine residues 108 and 110 replaced with alanine; elp3-2 with cysteine residues 118 and 121 replaced with alanine; elp3-4 with glycine 168 replaced with an arginine; elp3-3 with glycine 180 and 181 replaced with arginine. Two tyrosine residues Tyr 540 and 541 located at HAT domain were also replaced with alanine (elp3-5). (B) Four of the five site-specific elp3Δ mutants reduce telomeric silencing. Plasmids expressing wild type or each of the elp3 mutants were transformed into the elp3Δ mutant cells and telomeric silencing was assayed as described in Figure 1. (C) All five elp3 mutants reduce silencing of the ADE2 gene at telomeres. The ADE2 gene was integrated at the HMR locus and used to assay silencing of each of the five site-specific elp3Δ mutants. (D) The site-specific elp3 mutant cells are also sensitive to HU. Ten fold serial dilutions of yeast cells expressing wild type or each of the five site-specific elp3 mutants were spotted onto different concentrations of HU media as indicated.