Abstract
Phosphatidyl-myo-inositol mannosides (PIMs) are key glycolipids of the mycobacterial cell envelope. They are considered not only essential structural components of the cell but also important molecules implicated in host-pathogen interactions. Although their chemical structures are well established, knowledge of the enzymes and sequential events leading to their biosynthesis is still incomplete. Here we show for the first time that although both mannosyltransferases PimA and PimB′ (MSMEG_4253) recognize phosphatidyl-myo-inositol (PI) as a lipid acceptor, PimA specifically catalyzes the transfer of a Manp residue to the 2-position of the myo-inositol ring of PI, whereas PimB′ exclusively transfers to the 6-position. Moreover, whereas PimB′ can catalyze the transfer of a Manp residue onto the PI-monomannoside (PIM1) product of PimA, PimA is unable in vitro to transfer Manp onto the PIM1 product of PimB′. Further assays using membranes from Mycobacterium smegmatis and purified PimA and PimB′ indicated that the acylation of the Manp residue transferred by PimA preferentially occurs after the second Manp residue has been added by PimB′. Importantly, genetic evidence is provided that pimB′ is an essential gene of M. smegmatis. Altogether, our results support a model wherein Ac1PIM2, a major form of PIMs produced by mycobacteria, arises from the consecutive action of PimA, followed by PimB′, and finally the acyltransferase MSMEG_2934. The essentiality of these three enzymes emphasizes the interest of novel anti-tuberculosis drugs targeting the initial steps of PIM biosynthesis.
PIMs3 are unique mannolipids found in abundant quantities in the inner and outer membranes of the cell envelope of Mycobacterium spp. and a few other actinomycetes.4 They are based on a phosphatidyl-myo-inositol (PI) lipid anchor carrying one to six Manp residues and up to four acyl chains (for review see Refs. 1, 2). Based on a conserved mannosyl-PI anchor, they are also thought to be the precursors of the two major mycobacterial lipoglycans, lipomannan (LM) and lipoarabinomannan (LAM) (1, 2). PIMs, LM, and LAM are considered not only essential structural components of the mycobacterial cell envelope (3–6), but also important molecules implicated in host-pathogen interactions in the course of tuberculosis and leprosy (1).
Although the chemical structure of PIMs is now well established, knowledge of the enzymes and sequential events leading to their biosynthesis is still fragmentary. According to the currently accepted model, the biosynthetic pathway is initiated by the transfer of two Manp residues and a fatty acyl chain to PI in the cytoplasmic leaflet of the plasma membrane. Based on genetic and biochemical evidence, Korduláková et al. (5) identified PimA (MSMEG_2935 in Mycobacterium smegmatis mc2155) as the enzyme that catalyzes the first mannosylation step of the pathway transferring a Manp residue most likely to the 2-position of the myo-inositol (myo-Ins) ring of PI. In contrast, the identity of PimB′, the enzyme responsible for the transfer of the second Manp to the 6-position of the myo-Ins ring of PIM1, still remains controversial. The Rv0557 protein of Mycobacterium tuberculosis H37Rv (PimB; MSMEG_1113 in M. smegmatis mc2155) was originally characterized as PimB′ (7). However, the lack of an Rv0557 ortholog in the genome of Mycobacterium leprae and the fact that the disruption of this gene in M. tuberculosis Erdman did not significantly affect the biosynthesis of PIMs suggest that compensatory activities exist in the bacterium or that Rv0557 serves another primary function (8, 9). Somewhat supporting the latter hypothesis, the ortholog of Rv0557 in Corynebacterium glutamicum (NCgl0452, renamed mgtA) was implicated in the mannosylation of a novel glycolipid (1,2-di-O-C16/C18:1-(α-d-mannosyl)-(1→4)-(α-d-glucopyranosyluronic acid)-(1→3)-glycerol), and Rv0557 from M. tuberculosis was reported to functionally complement for this enzyme in a C. glutamicum knock-out mutant (10). However, to our knowledge this mannosylated glycolipid has never been reported in mycobacteria, and it remains unclear whether PimB serves a similar physiological function in Mycobacterium spp.
More recently, Lea-Smith et al. (11) have shown that the biosynthesis of Ac1PIM2 from Ac1PIM1 in C. glutamicum is catalyzed by NCgl2106 (Cg-PimB′). Disruption of the NCgl2106 gene totally abolished Ac1PIM2 production in the mutant, arguing against the existence of a compensatory activity associated with the corynebacterial PimB enzyme. Although Ac1PIM2 production in Cg-pimB′ and Cg-pimB′/Cg-pimB knock-out mutants was restored upon complementation with the M. tuberculosis Rv2188c gene (11, 12), direct evidence that Rv2188c carried out the same physiological function in mycobacteria has been lacking. Moreover, in light of the recent work by Torrelles et al. (9) showing an involvement of pimB (Rv0557) in the synthesis of LM and LAM in M. tuberculosis Erdman and of the demonstrated relaxed substrate specificity of the M. tuberculosis PimB (Rv0557) and PimB′ (Rv2188c) enzymes expressed in C. glutamicum (12), whether or not pimB and pimB′ could compensate for one another in mycobacteria remained open to speculation.
Both PIM1 and PIM2 can be acylated with palmitate at position 6 of the Manp residue transferred by PimA by the acyltransferase MSMEG_2934 (orthologous to Rv2611c from M. tb) to form Ac1PIM1 and Ac1PIM2, respectively (13). Ac1PIM2 can further be acylated at position 3 of the myo-Ins ring by an as yet unidentified acyltransferase to yield Ac2PIM2. Importantly, Ac1PIM2 and Ac2PIM2 are among the most abundant forms of PIMs found in mycobacteria and are considered both metabolic end products and intermediates in the biosynthesis of more polar forms of PIMs (PIM5 and PIM6), LM, and LAM.
In this work, clear evidence is provided that PimB′ (MSMEG_4253 in M. smegmatis mc2155) is the α-ManT responsible for the biosynthesis of PIM2 from PIM1 in mycobacteria and that no other ManT can compensate for a deficiency in this enzyme in M. smegmatis. Like PimA (5), PimB′ is essential to the growth of M. smegmatis. Cell-free assays using purified PimA and PimB′ and M. smegmatis membrane preparations provide new insights into the sequential events leading to the synthesis of the early forms of PIMs in mycobacteria.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of PimB′ from M. smegmatis in Escherichia coli
The M. smegmatis pimB′ gene (MspimB′, MSMEG_4253, 72% amino acid identity to Rv2188c) was amplified from genomic M. smegmatis mc2155 DNA by standard PCR using oligonucleotide primers pimB′_NdeI_Fwd (5′-GGAATTCCATATGACCCGGGTGTTGTTGGTCACC-3′, pimB′_ XhoI_Rev (5′-CCGCTCGAGCGCCTGACGCGCCTCGCGTCGG-3′), and Phusion DNA Polymerase (New England Biolabs). The PCR fragment was digested with NdeI and XhoI and ligated to the corresponding restriction sites of pET29a (Novagen) generating pET29a-MspimB′. The recombinant MsPimB′ protein (385 residues) has an additional peptide of eight amino acids (386LEHHHHHH393) at the C terminus that includes a histidine tag.
E. coli BL21(DE3)pLysS cells transformed with pET29a-MspimB′ were grown in 2× YT medium supplemented with 25 μg ml−1 kanamycin and 34 μg ml−1 chloramphenicol at 37 °C. MspimB′ expression was induced by adding 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (MP Biomedicals). After 4 h at 37 °C, cells were harvested and resuspended in solution A (50 mm Tris-HCl, pH 8.0) containing protease inhibitors (Complete EDTA-free, Roche Applied Science). Cells were disrupted by sonication (five cycles of 1 min), and the suspension was centrifuged for 20 min at 10,000 × g. The supernatant was applied to a HisTrap chelating column (1 ml; GE Healthcare) equilibrated with solution B (50 mm Tris-HCl, pH 8.0, 500 mm NaCl). The column was then washed with solution B until no absorbance at 280 nm was detected. Elution was performed with a linear gradient of 0–500 mm imidazole in solution B at 1 ml min−1. The resulting preparation displayed a single protein band when run on a 10% SDS-polyacrylamide gel stained with Coomassie Brilliant Blue (supplemental Fig. 1S). The purified enzyme was concentrated to 10 mg ml−1 in solution A containing 20% glycerol and stored at −80 °C until further use in enzyme assays.
Enzyme Assays
The enzymatic activity of MsPimA and MsPimB′ was monitored using a radiometric assay. The reaction mixture contained 0.0625 μCi of GDP-[C14]Man (specific activity, 305 mCi mmol−1; Amersham Biosciences), 10 μg of PI (Avanti Polar Lipids; liver PI, [M − H]−, m/z = 885.53, where the predominant species contains one polyunsaturated C20 and one C18 fatty acyl chain), 50 μg of purified MsPimA, MsPimB′, or a mix of MsPimA and MsPimB′ and 50 mm Tris-HCl, pH 7.5, in a final volume of 250 μl. In some assays, membrane preparations from M. smegmatis mc2155 (0.5 mg of proteins) served as the source of lipid acceptors. Reactions were incubated for 2 h at 37 °C and stopped with 1.5 ml of CHCl3/CH3OH (2:1, by volume). The PIM-containing organic phase was prepared and analyzed by TLC as described by Korduláková et al. (5). MsPimA was purified as described previously (14).
For structural analyses, 500 μm cold GDP-Man replaced GDP-[C14]Man in the assay mixture described above. The reactions were incubated overnight at 37 °C and stopped by adding 1.5 ml of CHCl3/CH3OH (2:1, by volume). The nonradioactive mannolipid products from 15 reactions were isolated by preparative TLC as described (5).
Matrix-assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry
Compounds 1–5 were mixed with an equal volume of matrix (2,5-dihydroxylbenzoic acid dissolved in 10 mg ml−1 acetonitrile/water, 50:50, 0.1% trifluoroacetic acid), and the molecular mass was measured in the negative ion mode by MALDI-TOF MS on a Bruker Ultraflex TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA). External calibration was performed using an eight component calibration mixture on a spot adjacent to the sample.
NMR Analysis
One-dimensional and two-dimensional NMR experiments were carried out at 25 °C in a Varian Inova 500-MHz NMR spectrometer (Varian Inc., Palo Alto, CA) using an HCN probe head equipped with shielded z-gradient. Samples were dissolved in 0.6 ml of CHCl3/CD3OD (2:1, by volume) and spectra acquired using a 5-mm NMR probe. Typical parameters used for one-dimensional 1H experiments were as follows: sweep width, 5500Hz; flip angle, 45°; time domain data points, 32,768; number of transients, 32 or 256; and relaxation delay, 1.5 s. For the complete structural analysis of PIM1, PIM2, and Ac1PIM2, two-dimensional experiments, including gradient-selected correlation spectroscopy, total correlation spectroscopy, heteronuclear single quantum coherence spectroscopy, and heteronuclear multiple bond correlation spectroscopy were carried out. Parameters used for two-dimensional correlation spectroscopy and total correlation spectroscopy were as follows: sweep width, 5500 Hz in both F2 and F1 dimensions; time domain data points, 2048; number of free induction decay with t1 increment, 512; number of transients, 32 or 256; and relaxation delay, 1.5s. Parameters used for heteronuclear single quantum coherence and heteronuclear single quantum coherence spectroscopy were as follows: sweep width, 5500 Hz in F2 and 30,188 Hz in F1; time domain data points, 2048; number of free induction decays with t1 increment, 256; number of transients, 32 or 256; and relaxation delay, 1.5 s. The acquired NMR data were processed using the TOPSPIN 2.1 software (Bruker GmbH, Karlsruhe, Germany).
Construction of M. smegmatis MspimB′ Conditional Mutant
Essentially the same strategy was used to construct a conditional MspimB′ mutant of M. smegmatis as was used earlier to generate an MspimA conditional mutant (5). The M. smegmatis MspimB′ gene (MSMEG_4253) and flanking regions were amplified from genomic M. smegmatis mc2155 DNA by standard PCR strategies using oligonucleotide primers MspimB′_KO_ApaI_fwd (5′-ATAATGGGCCCGCAAAACTGCGTGACCTGTACG-3′) and MspimB′_KO_SpeI_rev (5′-ATTATACTAGTGACCTCGGCGCCATCGACG-3′), and Phusion DNA polymerase (New England Biolabs). A disrupted allele of MspimB′, MspimB′::km, was constructed by cloning the kanamycin resistance cassette from pUC4K (GE Healthcare) into the AgeI and StuI sites of MspimB′, generating a 363-bp deletion within the coding sequence of MspimB′. MspimB′::km was then ligated to pJQ200xylE to yield pJQMspimB′KX, the vector used to achieve allelic replacement at the MspimB′ locus (5). The temperature-sensitive pCG76 derivative (15), pCGMspimB′, was used as the rescue plasmid to carry a functional copy of the MspimB′ gene in the conditional mutant.
Homology Modeling of MsPimB′
Homology modeling of MsPimB′ was performed with MODELLER 9 Version 4 (16) using the atomic coordinates of MsPimA complexed with GDP-Man (Protein Data Bank code 2GEJ (17)) as a template. Sequence alignment was carried out manually to match functionally conserved residues, predicted secondary structures, and hydrophobicity profiles. Secondary structures were predicted using the Jpred program (18). The models were assessed by the VERIFY_3D program.
RESULTS AND DISCUSSION
MsPimB′ Catalyzes in Vitro the Transfer of a Manp Residue to the 6-Position of the Myo-Ins Ring of PI
With the goal of determining the function of the mycobacterial PimB′ enzyme, a recombinant form of the M. smegmatis protein (MsPimB′) with a C-terminal histidine tag was produced in E. coli BL21(DE3)pLysS and purified to near homogeneity (supplemental Fig. 1S). As had been the case with the M. tuberculosis PimA protein earlier (5, 14, 17), attempts to produce the PimB′ enzyme from M. tuberculosis yielded relatively small amounts of soluble protein compared with the M. smegmatis version, and these efforts were thus not pursued further.
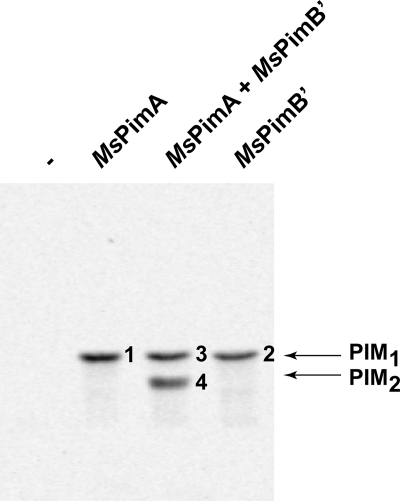
ManT assays were then run using different combinations of the purified MsPimA and MsPimB′ enzymes. When commercial liver PI and GDP-[14C]Man served as the acceptor and donor substrates in the assay, purified MsPimA (14) catalyzed the formation of PIM1 (mannolipid 1, Fig. 1). Unexpectedly, the formation of a 14C-labeled mannolipid with an Rf similar to that of PIM1 was also observed when purified MsPimB′ was used as the source of enzyme in the assay (mannolipid 2, Fig. 1). To further characterize mannolipids 1 and 2, nonradioactive products were purified by preparative TLC from reaction mixtures in which cold GDP-Man replaced GDP-[C14]Man. MALDI-TOF-MS analyses in the negative ion mode confirmed compounds 1 ([M − H]−, m/z = 1047.60) and 2 ([M − H]−, m/z = 1047.65) as PIM1 molecules (the [M − H]− m/z value of the commercial liver PI is 885.53) (supplemental Fig. 2S). MsPimB′ thus has the ability in vitro to transfer Manp from GDP-Man onto PI, generating PIM1.
FIGURE 1.
In vitro synthesis of PIM1 and PIM2 by purified recombinant forms of MsPimA and MsPimB′. TLC autoradiograph of reactions performed with purified recombinant MsPimA, MsPimB′, or a mix of MsPimA and MsPimB′ (1:1 by weight) using GDP-[C14]Man and commercial PI as the donor and acceptor substrates, respectively.
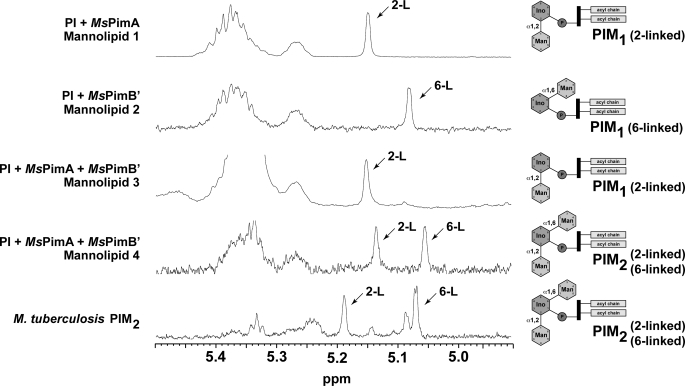
A combination of one-dimensional and two-dimensional NMR was then used to determine the position at which the Manp residues were attached to myo-Ins in mannolipids 1 and 2 (for details see supplemental material and supplemental Figs. 3S–6S) (19–21). As depicted in Fig. 2, the 1H NMR spectra of mannolipid 1 shows one peak at 5.14 ppm assigned to the α-anomeric proton of the Manp residue attached to position 2 of myo-Ins. The 1H and 13C chemical shift values of mannolipid 3 is exactly comparable with that of mannolipid 1, and therefore compound 3 was also assigned to 2-linked PIM1. In the spectra of compound 2, the peak at 5.072 ppm was assigned as the α-anomeric proton of the Manp residue attached to position 6 of myo-Ins. The 1H NMR spectra of mannolipid 4 shows two distinct peaks at 5.129 and 5.046 ppm assigned to α-anomeric protons of two Manp residues attached to the 2- and 6-positions of myo-Ins.
FIGURE 2.
NMR analysis of purified mannolipids. 1H NMR spectra of mannolipids 1–4. The 1H NMR spectra of mannolipid 1 shows one peak at 5.14 ppm assigned to the α-anomeric proton of the Manp residue attached to position 2 of myo-Ins. The 1H and 13C chemical shift values of mannolipid 3 are exactly comparable with that of mannolipid 1, and therefore compound 3 was also assigned to 2-linked PIM1. In the spectra of compound 2, the peak at 5.072 ppm was assigned as the α-anomeric proton of the Manp residue attached to position 6 of myo-Ins. The 1H NMR spectra of mannolipid 4 shows two distinct peaks at 5.129 and 5.046 ppm assigned to α-anomeric protons of two Manp residues attached to distinct positions of myo-Ins. Based on the combined two-dimensional NMR spectral analyses, the α-anomeric protons at 5.129 and 5.046 ppm are assigned to the peaks that are 2- and 6-linked to myo-Ins, respectively, and therefore, compound 4 is assigned to 2,6-linked PIM2 (see supplemental material and supplemental Figs. 3S–6S).
For the first time, direct evidence arising from the use of purified enzymes was thus provided that MsPimA catalyzes the transfer of a Manp residue from GDP-Manp to the 2-position of the myo-Ins ring of PI, and MsPimB′ catalyzes the transfer of a Manp residue to the 6-position.
Sequential Order of the Mannosylation Reactions Leading to the Formation of PIM2 from PI and GDP-Man
The simultaneous addition of purified MsPimA and MsPimB′ (1:1, w/w) to the reaction mixture described above yielded two products, mannolipid 3 and mannolipid 4 (Fig. 1). MALDI-TOF-MS analyses in the negative ion mode confirmed compound 3 ([M − H]−, m/z = 1047.60) as PIM1 and compound 4 as PIM2 ([M − H]−, m/z = 1209.71) (supplemental Fig. 2S). From this experiment, it can thus be concluded that MsPimA and MsPimB′ are sufficient for the formation of PIM2 from PI and GDP-Man to occur. The fact that no PIM3 or more mannosylated products were formed in the reaction even after prolonged incubation times further indicated that MsPimA and MsPimB′ are unable to mannosylate PIM products beyond PIM2. Thus MsPimA and MsPimB′ appear to each catalyze the transfer of one single Manp residue.
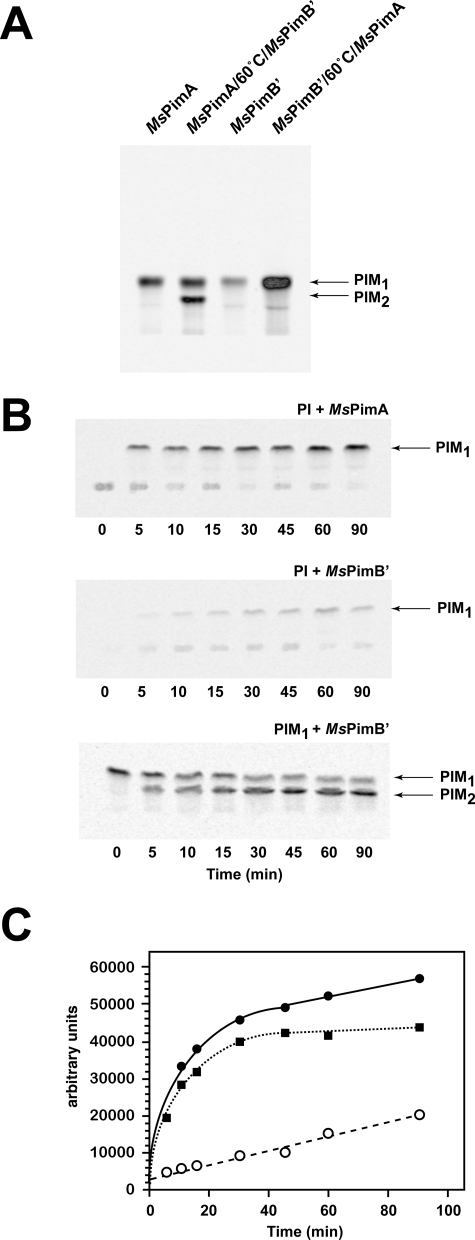
To determine the sequence of the reactions leading to the formation of PIM2, two independent assays were carried out in which purified MsPimA and MsPimB′ were added sequentially to the reaction mixture. In one of the assays, MsPimA was added first to a reaction mixture containing PI and GDP-[C14]Man. After 2 h of incubation, one-half of the reaction was stopped by the addition of CHCl3/CH3OH (2:1), and the other half was incubated at 60 °C for 15 min to inactivate the enzyme. Purified MsPimB′ was then added to the heat-inactivated assay mixture, and the reaction allowed to proceed overnight at 37 °C. In the second assay, MsPimB′ was added first to the reaction mixture, then inactivated as described above, and MsPimA finally added. That both MsPimA and MsPimB′ were inactivated by heat treatment was verified by running independent assays with each of the purified enzymes (supplemental Fig. 7S). Consistent with our previous results, both MsPimA and MsPimB′ catalyzed the transfer of a Manp residue from GDP-Man to PI to form PIM1 (Fig. 3A). The subsequent addition of MsPimB′ to the MsPimA reaction mixture clearly led to the synthesis of 14C-labeled PIM2 (Fig. 3A). In striking contrast, the addition of MsPimA to the MsPimB′ reaction mixture only resulted in the stimulation of PIM1 production with no detectable formation of PIM2 (Fig. 3A). We thus conclude from this experiment that although MsPimB′ recognizes the PIM1 product of MsPimA (with an α-1,2-linked Manp residue on the myo-Ins ring; Fig. 2) as an acceptor substrate, MsPimA is unable to transfer a Manp residue onto a PIM1 product bearing an α-1,6-linked Manp residue.
FIGURE 3.
PIM2 is produced by the sequential addition of mannosyl residues to PI and PIM1 transferred by MsPimA and MsPimB′, respectively. A, TLC autoradiograph of enzymatic reactions performed with purified recombinant MsPimA and MsPimB′ added sequentially to the reaction mixture. GDP-[C14]Man and commercial PI served as the donor and acceptor substrates in these reactions (see text for details). B, time course of transfer of [C14]Manp from GDP-[C14]Man onto PI and PIM1 (carrying an α-1,2-linked Manp residue) by MsPimA and MsPimB′. C, quantification of the reaction products shown in B. Solid circles, PIM1 product of MsPimA; solid squares, PIM2 product of MsPimB′; open circles, PIM1 product of MsPimB′.
With the transfer of Manp from GDP-Man onto PI catalyzed by MsPimB′ occurring 53 times slower than both the MsPimA-dependent transfer of Manp onto PI and the MsPimB′-dependent addition of Manp onto PIM1 (Fig. 3, B and C), it is clear that the different activities of the two enzymes with PI and, subsequently, PIM1 acceptors dictate the order in which the mannosylation of PI and PIM is to occur under physiological conditions. In further support of this assumption, the PIM1 product (mannolipid 3, Fig. 1) formed in a competition assay, where equal amounts of MsPimA and MsPimB′ were used as enzyme sources, exclusively consisted of α-1,2-linked Manp residues, as opposed to the expected mixture of α-1,2- and α-1,6-linked Manp residues if both enzymes had transferred Manp onto PI with comparable efficiencies (Fig. 2).
MsPimB′ Stimulates the Production of Ac1PIM2 in M. Smegmatis Membrane Preparations
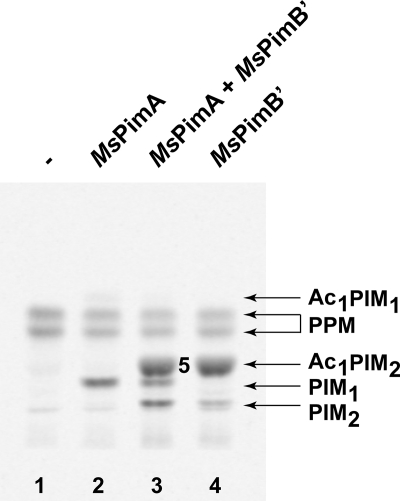
When membranes prepared from M. smegmatis mc2155 were used as a source of phospho(glyco)lipid acceptor, the addition of purified MsPimA clearly stimulated the synthesis of PIM1, accompanied by the accumulation of small amounts of Ac1PIM1 (Fig. 4, lane 2). The addition of MsPimB′ to the membrane preparations, in contrast, led to an even greater accumulation of a compound (mannolipid 5) with Rf properties similar to that of Ac1PIM2 (Fig. 4, lane 4). MALDI-TOF MS and NMR analyses confirmed the identity of this product as Ac1PIM2 containing two C16 and one C19 fatty acyl chains ([M − H]−, m/z = 1413.88) (supplemental Fig. 2S) (21), among which two fatty acyl chains are carried by the glycerol moiety and one acyl chain is attached to Manp residue located at position 2 of the myo-Ins ring (supplemental Fig. 8S). The acylation of the Manp residue transferred by PimA is thought to result from the action of the acyltransferase encoded by MSMEG_2934 in M. smegmatis mc2155 (13).
FIGURE 4.
Effects of adding purified MsPimA, MsPimB′, or both enzymes on the synthesis of PIMs by membrane preparations from M. smegmatis. Assays were carried out as described in the text using GDP-[C14]Man as the donor substrate and M. smegmatis membranes as a natural source of phospho(glyco)lipid acceptors. TLC autoradiograph of the reactions: lane 1, no purified enzyme was added; lane 2, purified MsPimA was added; lane 3, equal amounts of purified MsPimA and MsPimB′ were added simultaneously; lane 4, purified MsPimB′ was added.
Overall, the abundant de novo synthesis of Ac1PIM2 in the assay mixture containing purified MsPimB′ (Fig. 4, lane 4) suggests that significant amounts of PIM1 are available in the membranes of M. smegmatis or that the synthesis of this acceptor substrate is stimulated by the addition of purified MsPimB′ to the reaction mixture. This observation and the fact that radiolabeled Ac1PIM2 was on the contrary not detectable in the assay mixture in which only purified MsPimA was added (Fig. 4, lane 2) suggest that the physiological amounts of MsPimB′ present in the membranes of M. smegmatis may be rate-limiting in the formation of PIM2/Ac1PIM2. On the other hand, with almost all of the PIM2 product of MsPimB′ being instantly converted to Ac1PIM2 (Fig. 4, lane 4), the activity of the acyltransferase does not seem to be limiting in the membranes of M. smegmatis. In fact, saturation of this enzyme only became clearly visible when both purified MsPimA and MsPimB′ were added to the reaction mixture, resulting in the accumulation of abundant quantities of PIM1 and PIM2 (Fig. 4, lane 3). Finally, the quasi-exclusive occurrence of PIM2s under their acylated form (Ac1PIM2) in the assay where MsPimB′ was added (Fig. 4, lane 4), whereas the product of the reaction catalyzed by MsPimA essentially occurred as PIM1 (i.e. with no acylation on the Manp residue) (Fig. 4, lane 2), strongly suggests that the acyltransferase MSMEG_2934 preferentially acylates PIM2 over PIM1. Thus, despite MSMEG_2934 displaying acyltransferase activity on both PIM1 and PIM2 in vitro (13), it is likely that under physiological conditions the preferred pathway to Ac1PIM2 involves the transfer of both mannosyl residues onto PI prior to the acylation of the α-1,2-linked Manp residue.
Revised Model for the Early Steps of PIM Biosynthesis
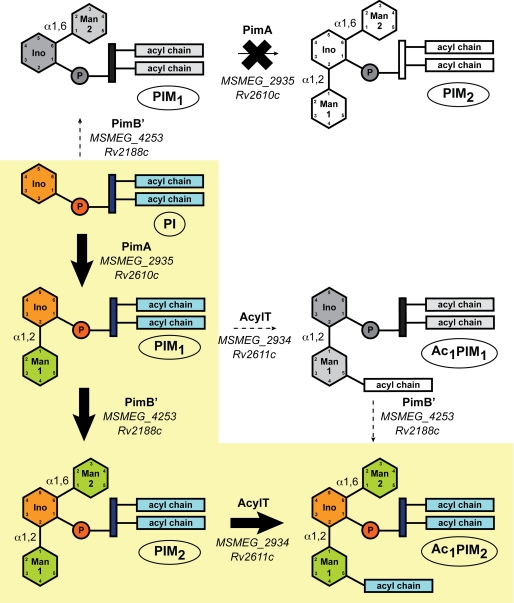
Based on present experimental evidence, a revised model for the early steps of PIM biosynthesis is presented in Fig. 5. As inferred from previous studies (5, 22–24) and now unambiguously demonstrated, MsPimA is the first enzyme engaged in the pathway. It is responsible for transferring a Manp residue from GDP-Man onto the 2-position of the myo-Ins ring of PI to form PIM1. MsPimB′ then transfers a second Manp residue from the same sugar donor to the 6-position of the myo-Ins ring of PIM1 yielding PIM2. Finally, the acyltransferase MSMEG_2934 acylates the Manp residue transferred by PimA to yield one of the major forms of PIM species found in mycobacteria, Ac1PIM2.
FIGURE 5.
Proposed pathway for the early steps of PIM biosynthesis in mycobacteria. The two pathways originally proposed for the biosynthesis of Ac1PIM2 in mycobacteria are shown. (i) PI is mannosylated to form PIM1. PIM1 is then mannosylated to PIM2, which is acylated to form Ac1PIM2. (ii) PIM1 is first acylated to Ac1PIM1 and then mannosylated to Ac1PIM2. Our experimental evidence indicates that although both pathways might co-exist in mycobacteria (13), the sequence of events PI → PIM1 → PIM2 → Ac1PIM2 is favored. As an important part of the literature concerning PIM studies refers to the nomenclature based on the M. tuberculosis H37Rv sequences, the Rv numbers of the proteins are also included.
MsPimB′ Is Essential for the Growth of M. smegmatis
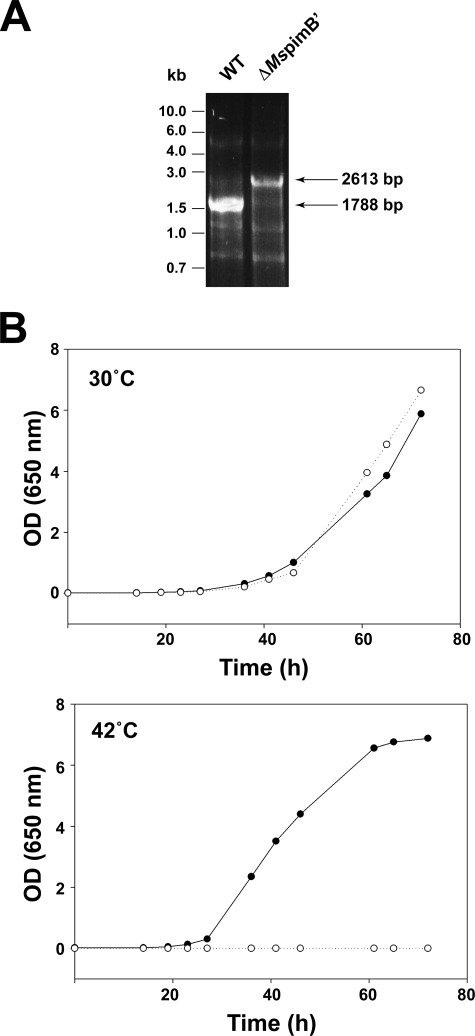
To investigate the essentiality or, on the contrary, possible redundancy of the ManT PimB′ in mycobacteria, a MspimB′ (MSMEG_4253) conditional mutant of M. smegmatis mc2155 was constructed. The methodology employed relies upon a suicide plasmid harboring the counter-selectable marker sacB to achieve allelic replacement, and a replicative temperature-sensitive plasmid (pCG76) to express a rescue copy of the gene of interest. Briefly, clones having undergone single crossover at the MspimB′ locus were first selected upon plating of mc2155/pJQMspimB′KX transformants on LB-Kan plates at 37 °C. Single crossover recombinants were grown in LB-Kan broth and then plated onto sucrose containing plates at 30 or 37 °C to select for allelic exchange mutants. No knock-out mutants were isolated at this stage strongly suggesting that MspimB′ was essential for growth regardless of the temperature used. To confirm this assumption, a conditional mutant of M. smegmatis was constructed. A temperature-sensitive rescue plasmid carrying a wild type copy of the MspimB′ gene, pCGMspimB′, was introduced in one of the single crossover recombinants, and the resulting merodiploids were plated onto LB-Kan-sucrose plates at 30 °C. Candidate conditional mutants were obtained in which allelic replacement at the chromosomal MspimB′ locus was confirmed by PCR (Fig. 6A). The conditional mutants grew normally at 30 °C in liquid broth or on plates, a temperature at which pCGMspimB′ replicates, but lost viability at 42 °C where the rescue plasmid is lost (Fig. 6B). Results thus indicated that MspimB′ is essential for the growth of M. smegmatis under the experimental conditions used. Therefore, despite the interchangeability of the M. tuberculosis PimB and PimB′ enzymes expressed in C. glutamicum in cell-free assays (12), the function of MsPimB′ cannot be compensated by any other ManTs, including MsPimB (MSMEG_1113; 75% identical to PimB from M. tuberculosis on a 375-residue overlap) in whole M. smegmatis cells.
FIGURE 6.
Essentiality of MspimB′ in M. smegmatis. A, PCR analysis showing allelic replacement at the MspimB′ locus. The wild type (WT) 1788-bp fragment is replaced by a 2613-bp fragment in the mutant because of the insertion of a 1.2-kb kanamycin resistance cassette. B, growth characteristics of the MspimB′ conditional mutant (○) and wild-type mc2155 parent strain (●) at 30 and 42 °C.
Structural Comparison of MsPimA and MsPimB′
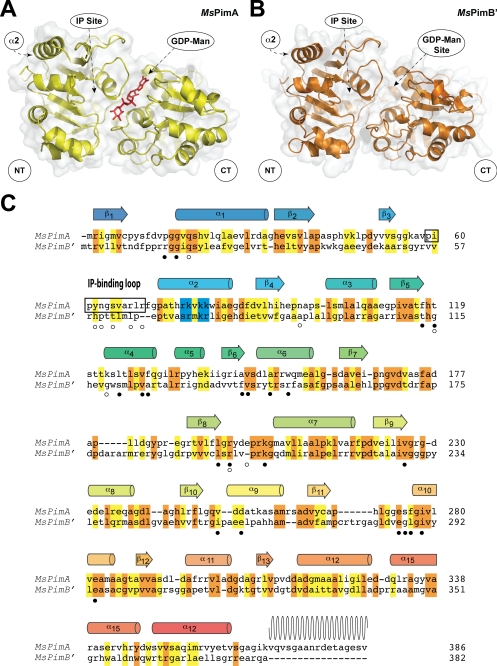
The α-ManTs MsPimA and MsPimB′ belong to the large GT4 family of glycosyltransferases, which includes more than 9800 proteins and at least 12 different enzymatic activities (see the Carbohydrate-Active enZymes data base). The GT4 family contains several enzymes of potential therapeutic significance and has been proposed as the ancestral “retaining” family from which enzymes with this type of stereochemistry have evolved (25, 26). MsPimA is one of the few GT4 enzymes whose three-dimensional structure has been solved. The enzyme displays the GT-B fold that consists of two Rossmann-like β-α-β domains separated by a large cleft that includes the catalytic center (Fig. 7A). The GDP-Man-binding site is located mainly in the C-terminal domain, where it makes a number of hydrogen bonds with the protein. Docking calculations and site-directed mutagenesis recently provided clear insights into the position of the polar head of the acceptor substrate, PI. Structural and enzymatic evidence support a model of interfacial catalysis in which MsPimA recognizes PI with its polar head within the catalytic cleft and the fatty acid moieties only partially sequestered from the bulk solvent. Membrane association is mediated by an interfacial binding surface in the N-terminal domain of the protein, which likely includes a cluster of basic residues in the amphipathic α-helix 2 (17) (Fig. 7A).
FIGURE 7.
Structural similarity between MsPimA and MsPimB′. A, experimental three-dimensional model of the crystal structure of MsPimA. B, three-dimensional homology model of MsPimA (Protein Data Bank code 2GEJ, see Ref. 17). C, structural alignment of MsPimA and MsPimB′. Secondary structure elements of the MsPimA three-dimensional structure are shown above the protein sequence. Wavy lines indicate disordered regions in the three-dimensional structure. The basic cluster in helix α2, which is proposed to be involved in membrane interaction, is highlighted in blue. Identical residues are shown in an orange background, and homologous residues are shown in a yellow background. Residues involved in the binding of GDP-Man and PI are denoted with solid and open circles, respectively.
A three-dimensional model of MsPimB′ was generated by homology modeling using the crystal structure of the MsPimA-GDP-Man complex as a template. Given that the two enzymes share only 28% overall sequence identity, the alignment was manually corrected incorporating information such as secondary structure prediction and conservation of functional residues. The overall predicted structure of MsPimB′ strongly resembles the experimental model of MsPimA (Fig. 7, A and B). Critical residues and their interactions are preserved in the two enzymes strongly supporting conserved catalytic and membrane association mechanisms (Fig. 7C). Two hydrophobic residues, Leu194 and Val226, that participate in the stabilization of the guanidyl heterocycle of GDP-Man in MsPimA are strictly conserved in MsPimB′ (Leu198 and Val229). Similarly, Val251 and Asp253, which confer nucleotide specificity to guanosine in MsPimA, are equivalent to Ile257 and Glu261, respectively, in MsPimB′. Gly16, Arg196, and Lys202, which are essential to stack the β-PO4 of GDP-Man in MsPimA, correspond to MsPimB′ residues Gly17, Arg200, and Lys205. Furthermore, Glu274 and His118, which are important for catalysis in MsPimA and several other GT-B enzymes, are equivalent to Glu286 and His114 in MsPimB′ (17). Interestingly, the MsPimB′ model predicts an amphipathic α-helix of the same length (14 residues) as the amphipathic α2 of MsPimA in which Arg78, Lys80, and Arg81 are also conserved. However, some of the key residues involved in PI binding, most notably the connecting loop between β3 and α2, differ between the two proteins reflecting their different acceptor substrate specificity. Overall, the structural conservation of MsPimA and MsPimB′ suggests that the two enzymes follow similar molecular mechanisms of substrate/membrane recognition and catalysis.
Concluding Remarks
Altogether, the results of our cell-free assays support a revised model for the early steps of PIM biosynthesis wherein the major PIM product of mycobacteria, Ac1PIM2, is formed via the sequential activity of PimA followed by PimB′ and, finally, the acyltransferase MSMEG_2934 (Fig. 5). Evidence is also provided for the first time that PimB′ is the ManT responsible for the addition of the Manp residue linked to position 6 of the myo-Ins moiety of PI in mycobacteria, and that a deficiency in its activity cannot be compensated by any other ManT of M. smegmatis. Thus, despite PimB and PimB′ having the potential to mannosylate the same substrates in in vitro assays (12), PimB and PimB′ clearly do not have redundant physiological functions in whole mycobacterial cells.
After PgsA1 (MSMEG_2933, Rv2612c in M. tuberculosis H37Rv), PimA (MSMEG_2935, Rv2610c in M. tuberculosis H37Rv), and the acyltransferase MSMEG_2934 (Rv2611c in M. tuberculosis H37Rv), PimB′ (Rv2188c in M. tuberculosis H37Rv) is now the fourth enzyme of the PIM pathway found to be essential in M. smegmatis and/or M. tuberculosis (5, 13, 27).5 Although this finding implies that PI, PIM1, and PIM2 are essential phospho(glyco)lipids, it is at present difficult to distinguish which of their roles as metabolic end products or as precursors for more mannosylated molecules (LM, LAM, and biosynthetic intermediates) specifically accounts for their essentiality. Ac1PIM2 appears to be a metabolic end product that accumulates at high steady state levels in the cells as well as a precursor for more polar forms of PIMs, LM and LAM. Both the PIM2 and the polar PIM contents of mycobacteria were found to directly impact on the permeability of the cell envelope (4, 5).6 Moreover, polar PIMs have been implicated in the homeostasis of the plasma membrane (6). In contrast to apolar PIMs, the essentiality of LM, LAM, and biosynthetic intermediates to the physiology of mycobacteria appears to depend on the Mycobacterium species. For instance, whereas the arabinosylation of LM was found to be essential to the growth of M. tuberculosis (28), this process is not essential to the viability of M. smegmatis (29). An M. smegmatis knock-out mutant defective in some aspects of the elongation of the mannan backbone of LM was also found to be viable, although its colonial morphology and growth rates were altered (30). Clearly, PIMs, LM, and LAM are likely to be involved in more than one critical function in mycobacterial cells, each of which or the combination of which might account for their essentiality. From a drug development perspective, the essential character of PIM biosynthetic enzymes and their relative restriction to mycobacteria and a few other actinomycetes emphasizes their interest as novel targets for anti-tuberculosis chemotherapeutic agents.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Karolin Luger (Department of Biochemistry and Molecular Biology, Colorado State University) for access to the protein purification facility, Dr. Hataichanok Scherman for help with plasmid constructs, and Dr. Jiang Zhang (Department of Microbiology, Immunology, and Pathology, Colorado State University) for help with NMR experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant AI064798 from NIAID.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text and Figs. 1S–8S.
PIM is used to describe the global family of phosphatidylinositol mannosides that carries one to four fatty acids (attached to the glycerol, inositol, and/or mannose) and one to six mannose residues. In AcxPIMy, x refers to the number of acyl groups esterified to available hydroxyls on the mannose or myo-inositol residues, and y refers to the number of mannose residues.
G. Stadthagen and M. Jackson, unpublished results.
N. Barilone and M. Jackson, unpublished results.
- PIM
- phosphatidyl-myo-inositol mannoside
- Manp
- mannopyranosyl
- ManT
- mannosyltransferase
- MALDI-TOF
- matrix-assisted laser desorption-ionization time-of-flight
- PI
- phosphatidyl-myo-inositol
- LM
- lipomannan
- LAM
- lipoarabinomannan
- myo-Ins
- myo-inositol.
REFERENCES
- 1.Gilleron M., Jackson M., Nigou J., Puzo G. (2008) in The Mycobacterial Cell Envelope ( Daffé M., Reyrat J. M. eds) pp. 75–105, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 2.Kaur D., Guerin M. E., Skovierova H., Jackson M. (2009) Adv. Appl. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goren M. B. (1984) in The Mycobacteria: A Sourcebook ( Kubica G. P., Wayne L. G. eds) Vol. 1, pp. 379–415, Marcel Dekker, Inc., New York [Google Scholar]
- 4.Parish T., Liu J., Nikaido H., Stoker N. G. (1997) J. Bacteriol. 179, 7827–7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korduláková J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B., Jackson M. (2002) J. Biol. Chem. 277, 31335–31344 [DOI] [PubMed] [Google Scholar]
- 6.Morita Y. S., Sena C. B., Waller R. F., Kurokawa K., Sernee M. F., Nakatani F., Haites R. E., Billman-Jacobe H., McConville M. J., Maeda Y., Kinoshita T. (2006) J. Biol. Chem. 281, 25143–25155 [DOI] [PubMed] [Google Scholar]
- 7.Schaeffer M. L., Khoo K. H., Besra G. S., Chatterjee D., Brennan P. J., Belisle J. T., Inamine J. M. (1999) J. Biol. Chem. 274, 31625–31631 [DOI] [PubMed] [Google Scholar]
- 8.Kremer L., Gurcha S. S., Bifani P., Hitchen P. G., Baulard A., Morris H. R., Dell A., Brennan P. J., Besra G. S. (2002) Biochem. J. 363, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torrelles J. B., Desjardin L. E., MacNeil J., Kaufman T. M., Kutzbach B., Knaup R., McCarthy T. R., Gurcha S. S., Besra G. S., Clegg S., Schlesinger L. S. (2009) Glycobiology 19, 743–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tatituri R. V., Illarionov P. A., Dover L. G., Nigou J., Gilleron M., Hitchen P., Krumbach K., Morris H. R., Spencer N., Dell A., Eggeling L., Besra G. S. (2007) J. Biol. Chem. 282, 4561–4572 [DOI] [PubMed] [Google Scholar]
- 11.Lea-Smith D. J., Martin K. L., Pyke J. S., Tull D., McConville M. J., Coppel R. L., Crellin P. K. (2008) J. Biol. Chem. 283, 6773–6782 [DOI] [PubMed] [Google Scholar]
- 12.Mishra A. K., Batt S., Krumbach K., Eggeling L., Besra G. S. (2009). J. Bacteriol. 191, 4465–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korduláková J., Gilleron M., Puzo G., Brennan P. J., Gicquel B., Mikusová K., Jackson M. (2003) J. Biol. Chem. 278, 36285–36295 [DOI] [PubMed] [Google Scholar]
- 14.Guerin M. E., Buschiazzo A., Korduláková J., Jackson M., Alzari P. M. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 518–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilhot C., Otal I., Van Rompaey I., Martìn C., Gicquel B. (1994) J. Bacteriol. 176, 535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 [DOI] [PubMed] [Google Scholar]
- 17.Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M. (2007) J. Biol. Chem. 282, 20705–20714 [DOI] [PubMed] [Google Scholar]
- 18.Cuff J. A., Barton G. J. (2000) Proteins Struct. Funct. Genet. 40, 502–511 [DOI] [PubMed] [Google Scholar]
- 19.Gilleron M., Nigou J., Cahuzac B., Puzo G. (1999) J. Mol. Biol. 285, 2147–2160 [DOI] [PubMed] [Google Scholar]
- 20.Gilleron M., Ronet C., Mempel M., Monsarrat B., Gachelin G., Puzo G. (2001) J. Biol. Chem. 276, 34896–34904 [DOI] [PubMed] [Google Scholar]
- 21.Gilleron M., Quesniaux V. F., Puzo G. (2003) J. Biol. Chem. 278, 29880–29889 [DOI] [PubMed] [Google Scholar]
- 22.Hill D. L., Ballou C. E. (1966) J. Biol. Chem. 241, 895–902 [PubMed] [Google Scholar]
- 23.Brennan P., Ballou C. E. (1967) J. Biol. Chem. 242, 3046–3056 [PubMed] [Google Scholar]
- 24.Takayama K., Goldman D. S. (1969) Biochim. Biophys. Acta 176, 196–198 [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Fleites C., Proctor M., Roberts S., Bolam D. N., Gilbert H. J., Davies G. J. (2006) Chem. Biol. 13, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 26.Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 27.Jackson M., Crick D. C., Brennan P. J. (2000) J. Biol. Chem. 275, 30092–30099 [DOI] [PubMed] [Google Scholar]
- 28.Goude R., Amin A. G., Chatterjee D., Parish T. (2008) J. Bacteriol. 190, 4335–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang N., Torrelles J. B., McNeil M. R., Escuyer V. E., Khoo K. H., Brennan P. J., Chatterjee D. (2003) Mol. Microbiol. 50, 69–76 [DOI] [PubMed] [Google Scholar]
- 30.Kaur D., McNeil M. R., Khoo K. H., Chatterjee D., Crick D. C., Jackson M., Brennan P. J. (2007) J. Biol. Chem. 282, 27133–27140 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.