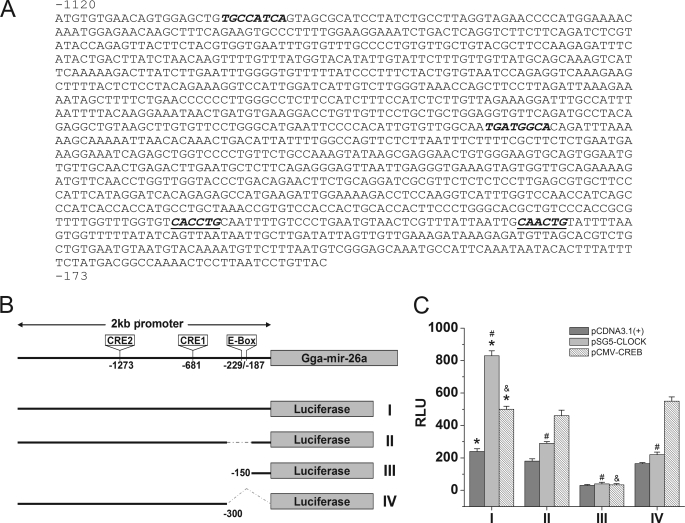
FIGURE 6.
CLOCK and CREB enhance gga-mir-26a expression in vitro. The luciferase reporter assays were performed by co-transfecting COS1 cells with one of the four reporter constructs (I, II, III, or IV) with either empty (pCDNA3.1), CLOCK (pSG5-CLOCK), or CREB (pCMV-CREB) expression vector. A, the sequence upstream of the gga-mir-26a promoter region (−1120 to −173) contained two CRE (italic type) and two E-box (italic and underlined type) cis elements. B, schematic diagram of the four luciferase reporter constructs of the gga-mir-26a promoter region: luciferase reporter with the 2-kb gga-mir-26a promoter region (I); 2-kb promoter region with a deletion mutation of E-box binding sites (Δ−330 to −150) (II); shorter promoter region with CRE and E-box deletion mutations (−150 to +1) (III); another E-box deletion mutation within the promoter region from −2 kb to −300 (IV). C, relative luminescence intensity (RLU) from the luciferase reporter assay after co-transfection with one of the four reporter constructs (I, II, III, or IV) and empty vector pCDNA3.1 (dark gray), pSG5-CLOCK (light gray), or pCMV-CREB (diagonal lines). Luciferase reporter assay results showed that CLOCK enhanced gga-mir-26a expression 3-fold, whereas CREB increased it 2-fold. Deletion of E-box (II, III, and IV) or both E-box and CRE (III) elements in the gga-mir-26a promoter region significantly reduced the transcriptional activities by CLOCK and CREB transcriptional factors. The empty vector pCDNA3.1 served as a control. n = 5 for each group. *, significant differences in RLU between pSG5-CLOCK or pCMV-CREB and pCDNA3.1 when co-transfected with full-length gga-mir-26a (I). #, significant decreases in RLU when pSG5-CLOCK is co-transfected with II, III, or IV compared with co-transfection with I. &, significant decrease in RLU when pCMV-CREB is co-transfected with III compared with co-transfection with I. p < 0.05.