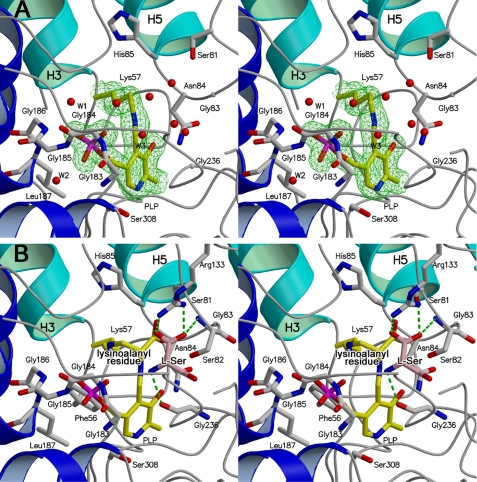
FIGURE 3.
Stereo view of the active site in spSR. The front and back of each figure show the solvent side (entrance of the active site) and the protein side (bottom of the active site), respectively. The side chains of the active site residues are depicted as stick models. The cofactor is shown in yellow with phosphate in red. The secondary structures of the small and large domains are drawn in light and deep blue, respectively, and the loops are drawn in gray. Water molecules are represented by red circles. A, close-up view of the active site of spSRw in the open form. A simulated annealing omit map contoured at the 1.0 σ level shows the PLP-Lys-57 internal Schiff base structure. The active site is exposed to the solvent and is filled with many water molecules. The water molecules numbered W1, W2, and W3 are conserved in spSRm in the closed form. B, close-up view of the spSRm complex with the substrate serine. PLP and the lysino-d-alanyl residue form a Schiff base. l-Serine (pink) is tentatively modeled into the active site. Arg-133, which is far from the active site in the open form, approaches the re-face side of PLP to form a salt bridge with the carboxylate of the substrate. The asparagine loop (Ser-81–Ser-82–Gly-83–Asn-84–His-85) moves to the active site center to recognize the carboxylate of the d-alanyl portion of the lysino-d-alanyl residue.