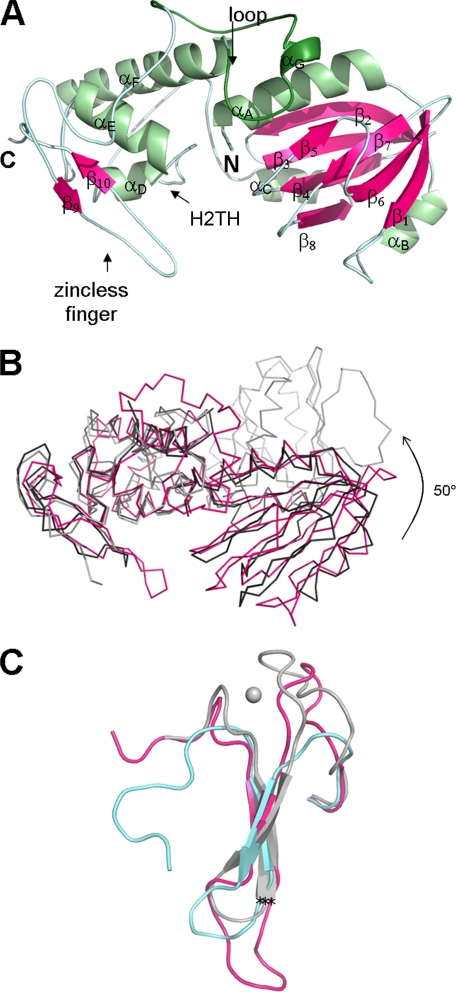
FIGURE 1.
A, ribbon diagram of unliganded MvNei1. The model from form II crystals comprises residues 2–289. The secondary structure elements were defined by DSSP (59) and are as follows: αA (4–18), β1 (22–27), αB (41–45), β2 (50–58), β3 (61–67), β4 (75–81), β5 (87–89), β6 (96–102), β7 (107–111), β8 (118–122), αC (125–133), αD (157–162), αE (173–183), αF (196–215), αG (226–229), β9 (265–267), and β10 (279–281). Helices are shown in light green and β-strands in magenta. The putative lesion binding loop is highlighted in a darker shade of green. B, comparison of MvNei1 with E. coli Nei (EcoNei). Superposition of MvNei1 (pink) with unliganded EcoNei (gray; PDB code 1Q3B (30) and the EcoNei trapped DNA complex (black; PDB code 1K3W (23)). C, close-up of the zinc-finger motif. Shown are residues 230–262 for EcoNei, 263–290 for hNEIL1, and 253–289 for MvNei1. The asterisks indicate the position of the Cα of the conserved arginine (Arg-252 in EcoNei and Arg-277 in hNEIL1 and MvNei1).