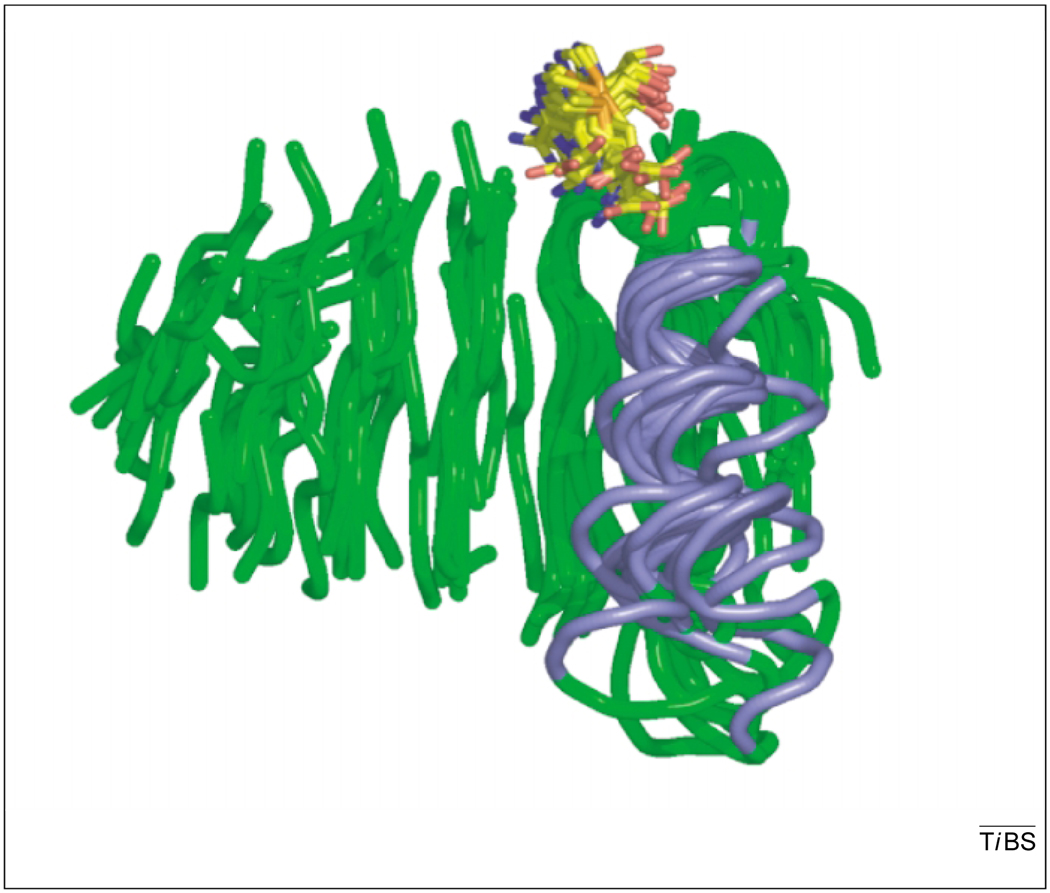
Fig. 2.
The strong structural similarity within the Class I methyltransferases (MTases) can been seen in the alignment of 14 structures via their β strands. For clarity, only the peptide backbone of the catalytic core containing the main β sheet (green) and the first α helix (blue) is shown. The cofactor (AdoMet or AdoHcy) is shown in stick model with carbon (yellow), oxygen (red), nitrogen (blue) and sulfur (orange). Structures include protein isoaspartate O-MTase (PDB code: 1I1N), protein arginine N-MTase (1F3L), cyclopropane-fatty-acyl-phospholipid synthase (1KPH), catechol O-MTase (1VID), M.TaqI (2ADM), M.HhaI (6MHT), CheR (1AF7), FtsJ (1EIZ), isoliquiritigenin O-MTase (1FP1), ErmC′ (1QAN), VP39 (1VPT), PrmC (1NV8), mj0882 (1DUS), hypothetical rv2118c (1I9G).