Abstract
Vibrio cholerae and many other marine and pathogenic bacteria posses a unique respiratory complex, the Na+-pumping NADH: quinone oxidoreductase (Na+-NQR)1, which pumps Na+ across the cell membrane using the energy released by the redox reaction between NADH and ubiquinone. In order to function as a selective sodium pump, Na+-NQR must contain structures that: 1) allow the sodium ion to pass through the hydrophobic core of the membrane, and 2) provide cation specificity to the translocation system. In other sodium transporting proteins, the structures that carry out these roles frequently include aspartate and glutamate residues. The negative charge of these residues facilitates binding and translocation of sodium. In this study we have analyzed mutants of acid residues located in the transmembrane helices of subunits B, D and E of Na+-NQR. The results are consistent with the participation of seven of these residues in the translocation process of sodium. Mutations at NqrB-D397, NqrD-D133 and NqrE-E95 produced a decrease of approximately ten times or more in the apparent affinity of the enzyme for sodium (Kmapp), which suggests that these residues may form part of a sodium-binding site. Mutation at other residues, including NqrB-E28, NqrB-E144, NqrB-E346 and NqrD-D88, had a large effect on the quinone reductase activity of the enzyme and its sodium sensitivity, but less effect on the apparent sodium affinity, consistent with a possible role in sodium conductance pathways.
The sodium pumping NADH:quinone oxidoreductase (Na+-NQR) is a unique prokaryotic respiratory enzyme capable of sustaining a sodium gradient across the plasma membrane, using the free energy released in the coupled oxidation of NADH and reduction of ubiquinone (1–3). Na+-NQR is composed of six subunits (NqrA-F) and contains five cofactors involved in the internal electron transfer: a non-covalently bound FAD and a 2Fe-2S center located in NqrF (4–8), two covalently-bound FMN’s in NqrB and NqrC (9–11), which have been shown to give rise to two anionic flavosemiquinone radicals, observed in the partially and fully reduced forms of the enzyme (12), and a non-covalently bound riboflavin molecule that is found as a stable neutral flavosemiquinone radical in the oxidized state of the enzyme (13, 14). This is notable because it is the only known instance in which riboflavin is present as a bona fide redox cofactor in any enzyme.
Topological analysis of the five membrane-bound subunits of Na+-NQR from Vibrio cholerae (NqrB-F) has revealed the presence of 17 acid residues in the membrane spanning segments of subunits B, D and E (15). Most of these acid residues are conserved across many of orthologs of the Na+-NQR family; and eight of these are also conserved in the related protein RNF, which has recently been proposed to also be a redox-driven sodium pump (16).
Ion transport by channels and pumps has at least two structural requirements. First, a substrate-binding site must provide a physiologically relevant affinity and specificity for transport. Second, a solute passageway, must allow the movement of ions through the hydrophobic core of the membrane. If the substrate-binding site is deeply buried in the membrane, passageways will be needed to connect the binding site to the two opposite sides of the membrane. Additionally, enzymes that function as ion pumps must be able to use an energy source to drive transport, and a gating mechanism must be in place to ensure the unidirectional movement of ions.
The occurrence of charged residues is unusual in hydrophobic environments of proteins, and represents only 1 to 3 % of the amino acid composition of transmembrane helices. This presumably reflects the large energy cost of introducing a charge into the hydrophobic environment. Thus, the presence of charged residues in the membrane, indicate an important role in protein function. In fact, these residues often form strong structural interactions between transmembrane segments, such as intramolecular helix-helix contacts or dimer associations (17). Alternatively, they may be parts of binding sites or pathways involved in movement of solutes through the membrane, in ion pumps or channels (18). In this respect, it has been proposed that these residues “prepay the Born energy” for the incorporation of ions into the membrane, which facilitates the transport process (18).
The identification of the sodium binding sites in proteins has been challenging, mainly because the different families of sodium transport proteins do not share conserved motifs. In many cases, the sodium binding residues are separated by segments of variable length in the primary sequence. In some cases, carbonyl oxygens of the peptide backbone provide the ligands for sodium binding, which allows much more sequence variability than if the binding sites, are in amino acid side chains. Moreover, water molecules can be ligands for sodium (19). The recognition of sodium binding sites is not trivial, even with the availability of crystallographic data, due to the similar characteristics of sodium atoms and water molecules. Nayal and Di Cera (20) studied this question by means of valence calculations on protein crystallographic structures and proposed an algorithm to distinguish bound sodium ions from internal water molecules. They concluded that an optimal sodium binding site consists of six oxygen atoms arranged in an octahedral geometry.
We have characterized mutants in which conserved glutamate and aspartate residues located in transmembrane helices of the Vibrio cholerae Na+-NQR were substituted by non-polar residues (A, G or L). Mutation of some of these residues led to significant decreases in the sodium-dependent quinone reductase activity (Qred), and thus in the coupled sodium pumping activity. We have identified seven acid residues, which appear to be of special importance for sodium translocation by the enzyme. Three of these mutations led to large decreases in the apparent affinity for sodium (Kmapp), consisting with a role as components of a sodium binding site. Although we cannot rule out the possibility that some of these mutations exert their effects indirectly through structural perturbation of the enzyme.
EXPERIMENTAL PROCEDURES
Mutant construction
Mutants were constructed with the Quikchange site-directed mutagenesis kit (Stratagene), using as template the wild-type nqr operon from Vibrio cholerae cloned into the pBAD vector (Invitrogen), as reported previously (6, 7). Primers were designed to replace the acid residues by small non-polar amino acids, such as A, G or L, and to simultaneously incorporate new restriction sites to aid in identification of mutants. Generally, Alanine residues were introduced but in some cases it was necessary to use Glycine or Leucine instead in order to create the ne restriction site. A list of the primers used in this study is found in Table 1. Mutations were verified by restriction digestion analysis and by direct sequencing of the plasmids. Mutant plasmids were introduced by electroporation into the Δnqr V. cholerae strain.
Table 1.
Primers designed for the site directed mutagenesis used in this work.
| Subunit | Mutation | Primer sequence (5’→3’) |
|---|---|---|
| B | ||
| E28A | GTGGTTTGCCCTGTATGCCGCGGCGGCGACGCTG | |
| E144L | CTATCTACGCTACGGTGTTCATCGTCGGTGGTTTCTGGCTCGTGTTG TTCTGTATGGTGC GCAAGCATGAAGTCAAC |
|
| D223A | GCACAGATCTCAGGAGCTCTAGTATGGACTGCGG | |
| E274A | GGTAACATCCCAGGTTCAATTGGCGCCGTGTCTACTCTGGCACTCA TGATTG |
|
| D346A | GGTATGTTCTTCATGGCGACGGCGCCATGGTCTGCGTCCTTCACC | |
| E380A | GTGAACCCGGCTTACCCAGCCGGCATGATGCTGGCGATC | |
| D397A | GCGAACCTATTTGCGCCACTGTTTGCGCATGTGGTTGTAGAGAAAT ATCAAG |
|
| D | ||
| D17A | GAGTGTGTTAGCCCCAGTGCTAGCCAACAACCCGATTGCACTGC | |
| E39L | GCGGTAACCACTAAGCTGTTAACGGCATTTGTTATGACGCTA | |
| D87L | TCGCTTCGTTAGTTATCGTGGTACTGCAGATTCTGAAAGCGTATCTG | |
| E119A | CGTAATGGGTCGTGCCGCGGCGTTTGCAATGAAGTCTG | |
| D133A | TCTGAACCAATTCCTTCTTTCATTGCCGGCATTGGTAACGGTTTAGG A |
|
| E153A | CTGATGACGGTTGGTTTCTTCCGAGCTCTTTTAGGCTCAGGTAAG | |
| E | ||
| E15A | CTGCTGGTGAAATCGATTTTCATCGCTAACATGGCACTAAGCTTCTT CCTAGGG |
|
| E95A | GCGGCATTATGACAGATTCTGGCTATGATCCTCGATCGC | |
| E138A | CAGCGCGACTACAGCTTTGCTGCTAGCGTGGTATACGGTTTCGGTT C |
|
| E162G | GTCGCTCTTGCAGGTATCCGCGGTAAGATGAAGTATTCAGAC |
The underlined bases indicate the sites that were modified to introduce the mutations
Cell culture
Δnqr V. cholerae cells containing the wild type or mutant nqr operon on the pBAD plasmid were grown at 37 °C in LB medium in New Brunswick BioFlo-5000 fermentors under constant aeration and agitation. Arabinose was added as the inducer of expression, as described previously (6, 7).
Protein purification
Protein purification was performed using Ni-NTA (Qiagen) affinity chromatography (6) and protein concentration was measured with the BCA assay (Pierce). Flavin content was measured spectrophotometrically at 450 nm (εFAD= 12.1 mM−1 cm−1) in 6 M Guanidine-HCl (6). In all the cases the flavin/protein molar ratio varied from 3.1 to 3.6, consistent with the presence of four flavin molecules in the enzyme. This argues against the possibility that some of the acid residue mutations affect the binding of flavin cofactors.
Activity measurements
Specific activities were measured spectrophotometrically as reported previously (6). Measurements were carried out in buffer (50 mM Tris-HCl, pH 8; 1mM EDTA; 5% (v/v) glycerol; 0.05% (w/v) n-dodecyl β-maltoside) containing 150 µM K2-NADH and 50 µM Q-1. The NADH dehydrogenase (NADH-DH) activity was measured at 340 nm (εNADH= 6.22 mM−1 cm−1). The NADH-dependent quinone reductase activity (Qred) was measured at 282 nm, using the molar absortivities for quinol (ε= 2.7 mM−1 cm−1) and quinone (ε= 14.5 mM−1 cm−1) in aqueous environment. The NADH oxidase (NADHox) activity was calculated as the difference between the NADH dehydrogenase and NADH-dependent quinone reductase activities, which presumably corresponds to the rate of electron branching to oxygen. In fact, under anaerobic conditions the rate of the NADH oxidase reaction decreased in more than 95% for wild-type enzyme (not shown).
Enzyme reconstitution
Wild-type and mutant enzymes were reconstituted into liposomes following the general recommendations of Rigaud et al. (21). Two mg of purified enzyme were added to 15 mg/ mL of a total E. coli phospholipid extract (Avanti lipids) in buffer containing 25 mM n-octyl glucoside (detergent/phospholipid molar ratio of 1.3), 100 mM HEPES, 150 mM KCl, 1 mM NaCl, 1 mM EDTA, pH 7.5. The detergent was slowly removed by adding small amounts of SM2 Biobeads (Bio-Rad), following the protocol described by Verkhovskaya et al. (22).
Formation of membrane potential (ΔΨ)
ΔΨ was measured spectrophotometrically at 625-minus-587 nm in the preparations of enzyme reconstituted in proteoliposomes, using Oxonol VI (23, 24). The reaction buffer contained 200 µM NADH, 100 µM Q-1, 100 mM NaCl, 100 mM HEPES, 150 mM KCl, 1 mM EDTA, pH 7.5. Calibration of the spectrophotometric signal of Oxonol VI was performed by varying the diffusion potential of potassium (23).
RESULTS
Topological studies on Na+-NQR from Vibrio cholerae have found 17 negatively charged amino acid residues in the transmembrane α-helices of subunits B, D and E, most of which are conserved across members of the Na+-NQR family (15). Many of these residues are also conserved in the related enzyme RNF, which has a similar cofactor and subunit composition to Na+-NQR. RNF has also been proposed to function as a redox driven sodium pump (16, 25). We compared the sequences of the transmembrane segments of Na+-NQR with the corresponding regions of the homologous subunits of RNF (Table 2). Eight of the seventeen acidic residues in Na+-NQR are completely conserved in RNF, while an additional six are semi-conserved. Conservation in the more distantly related RNF is an additional indication of the functional importance of these amino acid residues.
Table 2.
Sequence alignments of the transmembrane segments of the Na+-NQR subunits B, D and E, against the homologous subunits of RNF.
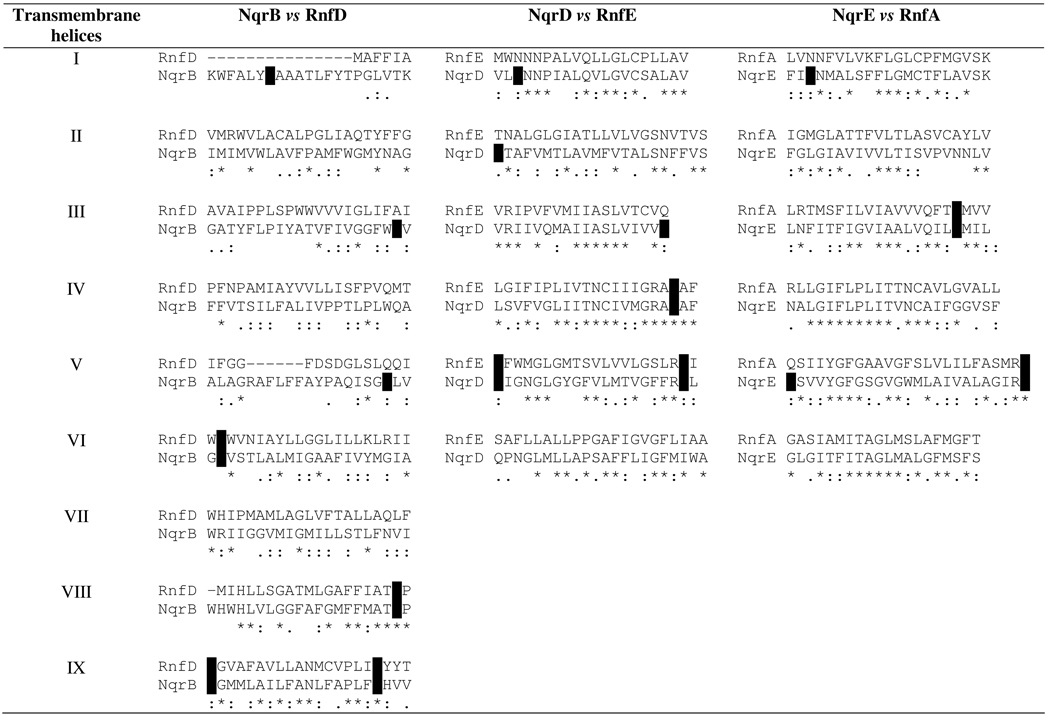 |
Conserved acidic residues are indicated in white on black. The transmembrane segments of each subunit were obtained with the algorithms used previously (15). In cases where a consensus transmembrane helix was not well defined, all possible amino acids were included in the alignment, resulting in some helices appear longer than others.
The NqrB subunit contains a total of seven acid residues, one in each of transmembrane helices I (E28), III (E144), V (D224), VI (E274) and VIII (D346), and two in helix IX (E380 and D397). Three of these are conserved in the corresponding Rnf subunit D (E274, D346 and D397); two are present as semi-conservative substitutions, (D224→Q and E380→D), and two are not conserved (E28 and E144). NqrD contains a total of six acid residues, one in each of transmembrane helices I (D17), II (E39), III (D87), and IV (E119), and two in helix V (D133 and E153). Three of these residues are conserved in the corresponding RnfE (E119, D133 and E153), two are present as semi-conservative substitutions (D17→N and D87→Q), and one (E39) is not conserved. NqrE contains a total of four acid residues, one in each of transmembrane helices I (E15) and III (E95) and two in helix V (E138 and E162). Two of these residues are conserved in the corresponding RnfA (E95 and E162) and the other two are present as semi-conservative substitutions (E15 →N and E138 →Q) (Table 2).
Thus, all but three of the acid groups in the transmembrane helices of Na+-NQR are either conserved in RNF or are present as semi-conservative substitutions, in which the new amino acid still contains an oxygen atom in the side chain (glutamine, asparagine or threonine). This suggests that a full or partial negative charge is required for the interactions in which these amino acids participate.
Activity measurements
We have analyzed the catalytic activity of wild-type Na+-NQR and the acid group mutants, and studied the effect of sodium concentration on these reactions. The physiological redox reaction of Na+-NQR is the transfer of electrons from NADH to quinone, which drives the pumping of sodium. This reaction can be measured by following either the oxidation of NADH at 340 nm (NADH dehydrogenase = NADHDH) or the reduction of quinone at 282 nm (Qred). However, as a side-reaction the enzyme can also donate electrons to dioxygen, producing superoxide (26–28) (NADH oxidase activity = NADHox). This side-reaction is often observed in the isolated enzyme, resulting in a rate of quinone reduction (Qred) that is lower than the rate of NADH consumption (NADHDH) in the presence of oxygen. Here, the NADHox activity was calculated from the difference between NADHDH and Qred activities, which is presumed to reflect the rate of electron leakage to oxygen.
In the absence of sodium the consumption of NADH by wild-type Na+-NQR was 6 to 7 times faster than the reduction of quinone, which indicates that 80–90% of the electrons from NADH flow to oxygen (NADHox/NADH-DH = 0.85) (Table 3). Addition of sodium produced a reproducible 20 to 40% increase in the NADHDH activity, but also led to a 6- to 8-fold increase in the rate of Qred and consequently, a large effect on the electron branching between the NADHox and Qred reactions, in which approximately 90% of the electrons go to the Qred activity (QredNa+ /NADH-DHNa+= 0.86).
Table 3.
Steady-state activities of wild–type and acid mutants of Na+-NQR in the presence and absence of saturating concentrations of sodium (100 mM NaCl).
| Conserved | no Na+ | Na+ | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| in RNF | |||||||||||
| Subunit | Mutant | NADH DH |
Q Red |
NADH Ox |
NADH- DH |
Q Red | NADH Ox |
QredNa+/Qred | |||
| WT | 437.3 | 65.4* | 374.9 | 557.7 | 521.4* | 74.1 | 8.0 | ||||
| B | |||||||||||
| E28A | 370.0 | 37.2 | 332.8 | 474.8 | 69.7 | 405.0 | 1.9 | ||||
| E144L | 431.0 | 46.4 | 384.6 | 529.1 | 115.0 | 414.1 | 2.5 | ||||
| D224A | : | 334.1 | 58.5 | 275.6 | 522.4 | 226.5 | 295.9 | 3.9 | |||
| E274A | * | 378.6 | 70.2 | 308.5 | 517.2 | 315.9 | 201.3 | 4.5 | |||
| D346A | * | 433.4 | 57.4+ | 406.3 | 535.8 | 66.3+ | 500.6 | 1.2 | |||
| E380A | : | 326.7 | 64.9 | 261.9 | 396.9 | 254.0 | 142.9 | 3.9 | |||
| D397A | * | 331.7 | 53.2+ | 312.4 | 366.8 | 58.9+ | 349.5 | 1.1 | |||
| D | |||||||||||
| D17A | : | 365.2 | 58.1 | 307.1 | 477.7 | 303.4 | 174.4 | 5.2 | |||
| E39L | 334.9 | 60.4 | 274.5 | 417.2 | 244.9 | 172.3 | 4.1 | ||||
| D88L | : | 477.8 | 31.5 | 446.3 | 533.6 | 58.9 | 474.7 | 1.9 | |||
| E119A | * | 365.6 | 54.2 | 311.4 | 466.4 | 312.6 | 153.8 | 5.8 | |||
| D133A | * | 486.0 | 25.4 | 460.6 | 545.1 | 42.3 | 502.8 | 1.7 | |||
| E153A | * | 397.5 | 67.4 | 330.1 | 567.4 | 300.7 | 266.7 | 4.5 | |||
| E | |||||||||||
| E15A | : | 421.3 | 70.6 | 350.7 | 612.8 | 350.4 | 262.5 | 5.0 | |||
| E95A | * | 505.3 | 50.2+ | 474.3 | 544.8 | 69.6+ | 504.7 | 1.4 | |||
| E138A | : | 367.1 | 64.3 | 302.8 | 488.5 | 383.0 | 105.5 | 6.0 | |||
| E162G | * | 366.1 | 62.5 | 303.6 | 561.4 | 290.2 | 271.3 | 4.6 | |||
Steady-state activity determination of wild–type and Na+-NQR mutants was performed in the presence and absence of saturating concentrations of sodium (100 mM NaCl).
completely conserved in RNF; (: ) semiconserved. Activities were measured as described in Materials and Methods section.
NADH DH; NADH dehydrogenase activity, Q red; quinone reductase activity, NADH ox; NADH oxidase activity.
All activities are expressed in turnover rate (s−1).
The data indicated were obtained with enzyme that have been further purified by gel filtration chromatography (HiLoad 16/60 Superdex 200 column) using buffer containing 50 mM Tris-HCl, pH 8, 100 mM NaCl, 1 mM EDTA, 0.1% (w/v) n-dodecyl β-maltoside, 5% (v/v) glycerol.
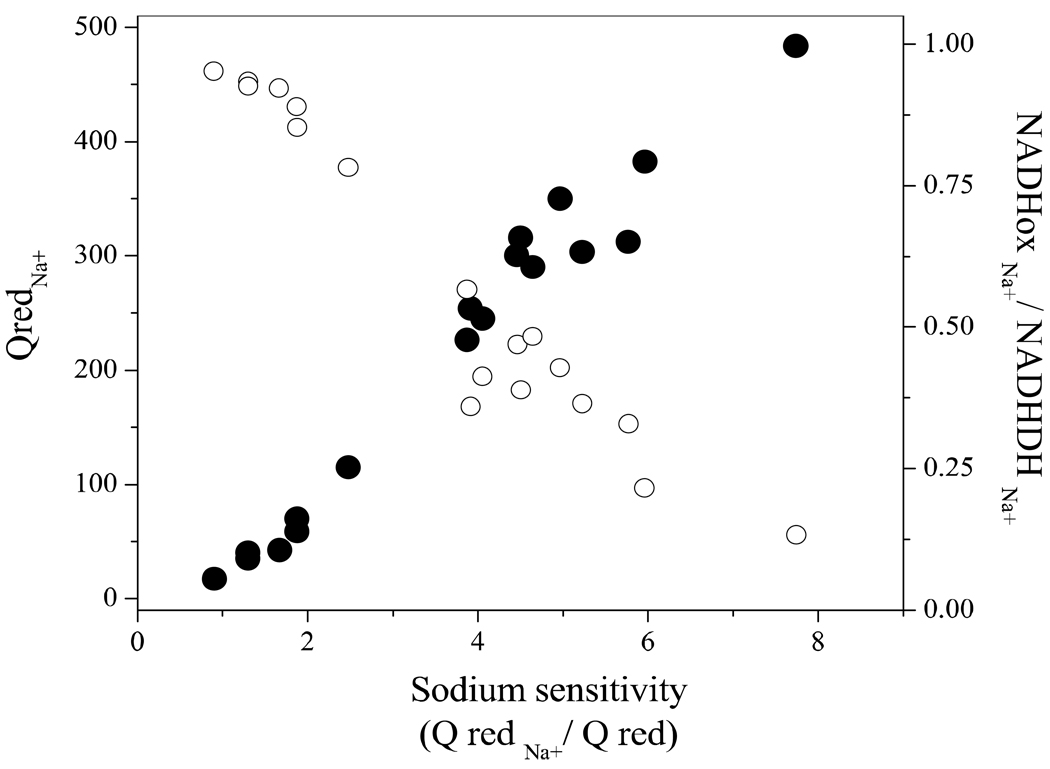
Table 3 shows the results of activity measurements on wild-type Na+-NQR and the acid group mutants in the presence and absence of sodium. In the wild-type enzyme sodium accelerates Qred activity 7–8 fold. This effect was attenuated in the acid mutants. In some mutants such as NqrB-D397, NqrB-D346A and NqrE-E95A the effect of sodium on Qred activity was almost eliminated, while in other mutants a smaller effect was observed. Figure 1 shows the relationship between the sodium sensitivity of the Qred activity (QredNa+/Qred), the net Qred activity and the fraction of electrons from NADH that flow to oxygen. Mutants with the least sodium sensitivity showed both the lowest Qred activity and the largest electron leak to oxygen (in the presence of sodium). These results may be expected if the acid group mutants restricted the access of sodium to the enzyme.
Figure 1.
Correlation between the sodium sensitivity (QredNa+/ Qred) of the wild-type and mutant Na+-NQR with the Q reductase activity (QredNa+; ●), and the fraction of electrons leaking to oxygen (NADHoxNa+/ NADH-DHNa+; ○) in the presence of sodium.
Some of the mutants (especially those with the least sodium sensitivity) displayed an apparent decrease in activity, compared to wild type, even in the absence of sodium. Since the determination of the concentration of enzyme was performed spectrophotometrically, it was possible that contaminants in some mutant preparations might have increased the apparent concentration of enzyme in the sample and therefore have decreased the apparent specific activity. To check for this possibility, the mutants NqrB-D346, NqrB-D397A and NqrE-E95A were further purified by gel filtration chromatography. As shown in Table 3, these purified mutant enzymes had Qred activity values very similar to wild-type Na+-NQR, in the absence of sodium (data marked by asterisks). These results are consistent with the idea that these mutants exert their major effect by perturbing sodium transport in the enzyme.
We also studied the ion selectivity, comparing the effects of sodium and potassium on the activity of Na+-NQR wild type and mutant enzymes (Table 4). The results with the wild type enzyme confirm earlier findings by Hayashi and Unemoto (26) using the enzyme from Vibrio alginolyticus (see also (6, 27, 29). The effect of sodium on the Qred activity is approximately seven times larger than the effect of potassium, indicating that the dominant effect is not due to ionic strength. However, the apparent stimulation of the NADHDH and NADHox activities may be due to ionic strength, since the effects of sodium and potassium are similar.
Table 4.
Effect of sodium and potassium on the activity of wild–type and acid mutants of Na+-NQR.
| Activity with cation/ Activity without cation* | |||||||
|---|---|---|---|---|---|---|---|
| NADH DH | Qred | NADHox | |||||
| Subunit | Mutant | Na+ | K+ | Na+ | K+ | Na+ | K+ |
| WT | 1.31 | 1.18 | 7.84 | 1.02 | 0.58 | 0.91 | |
| B | |||||||
| E28A | 1.27 | 1.32 | 1.71 | 0.90 | 1.21 | 1.37 | |
| E144L | 1.30 | 1.32 | 2.66 | 0.96 | 1.00 | 1.37 | |
| D224A | 1.88 | 1.33 | 4.21 | 1.09 | 0.96 | 1.37 | |
| E274A | 1.78 | 1.42 | 5.2 | 0.85 | 0.84 | 1.53 | |
| D346A | 1.21 | 1.29 | 1.21 | 1.04 | 1.21 | 1.31 | |
| E380A | 1.47 | 1.26 | 3.8 | 0.87 | 0.79 | 1.32 | |
| D397A | 1.09 | 1.38 | 1.05 | 1.09 | 1.13 | 1.38 | |
| D | |||||||
| D17A | 1.96 | 1.40 | 8.75 | 1.11 | 0.93 | 1.44 | |
| E40L | 1.55 | 1.41 | 5.72 | 1.00 | 0.83 | 1.48 | |
| D88L | 1.09 | 1.15 | 1.54 | 0.94 | 1.04 | 1.18 | |
| E119A | 1.86 | 1.61 | 6.18 | 1.02 | 1.18 | 1.68 | |
| D133A | 1.25 | 1.42 | 1.89 | 0.95 | 1.18 | 1.48 | |
| E153A | 1.56 | 1.36 | 7.82 | 1.16 | 0.78 | 1.39 | |
| E | |||||||
| E15A | 1.98 | 1.37 | 7.57 | 1.10 | 1.19 | 1.41 | |
| E95A | 1.41 | 1.27 | 1.50 | 1.32 | 1.40 | 1.26 | |
| E138A | 1.75 | 1.31 | 8.26 | 1.08 | 0.58 | 1.35 | |
| E162G | 1.78 | 1.38 | 8.30 | 0.88 | 0.88 | 1.44 | |
Activities were measured as described under materials and methods in the absence of cations and in the presence of NaCl (100 mM) or KCl (100 mM).Activities are expressed in turnover rate (s−1).
The same pattern is generally observed in the mutants. In a few cases where there is very little activation by sodium, the difference between sodium and potassium activation is not large. However, in no case, does the mutation lead result in a notable increase in activation of Qred activity by potassium.
Sodium pumping activities
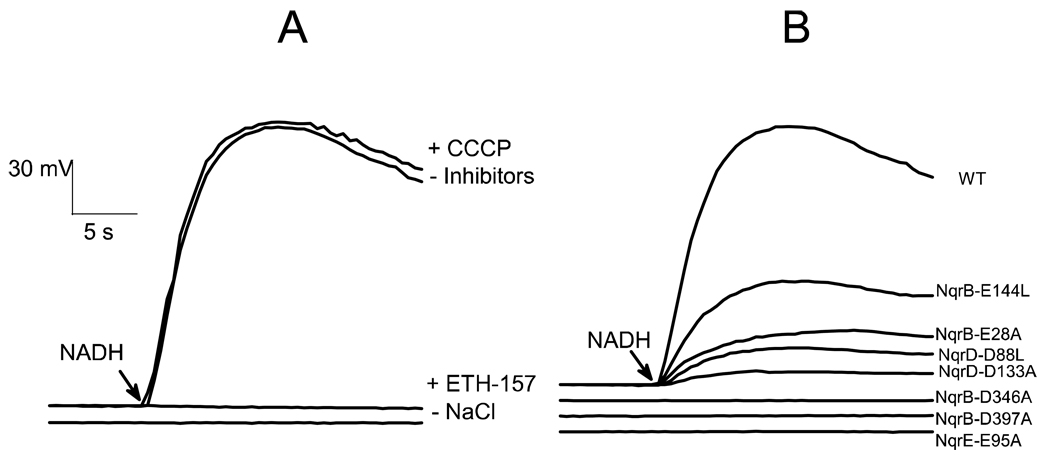
Sodium pumping activity was measured spectrophotometrically by means of the ΔΨ-sensitive probe Oxonol VI, which partitions across the membrane in a voltage sensitive manner (23, 24) (Table 5). In order to have a simple system that would allow us to address the activity of Na+-NQR directly, these measurements were made on the isolated enzyme reconstituted into phospholipid vesicles. To obtain a quantitative measurement, the spectrophotometric signal of Oxonol VI was calibrated by means of diffusion potential created with potassium and valinomycin. Figure 2A shows that the reconstituted wild-type enzyme is able to sustain a ΔΨ of about −160 mV, which is insensitive to the protonophore CCCP and is collapsed by the addition of the sodium ionophore ETH-157, indicating that the ΔΨ is the result of electrogenic sodium translocation. The results in Figure 2B show that the mutants NqrB-D346A, NqrB-D397A and NqrE-E95A, in which Qred activity is insensitive to sodium, were unable to form a sodium gradient. The mutants NqrB-E28A, NqrB-E144L, NqrD-D133A, and NqrD-D88L, in which sodium stimulation of Qred activity was lowered significantly, but not eliminated, produced diminished levels of ΔΨ.
Table 5.
ΔΨ formation in inverted vesicles and wild-type and mutant enzymes reconstituted in proteoliposomes.
| Na+ stimulation | ||||
|---|---|---|---|---|
| Subunit | Mutant | Purified Enzyme | Reconstituted Enzyme | ΔΨ (mV) |
| WT | 7.74 | 9.6 | −165 | |
| B | ||||
| E28A | 1.87 | 2.6 | −35 | |
| E144L | 2.48 | 3.5 | −65 | |
| D224A | 3.87 | −90 | ||
| E274A | 4.50 | |||
| D346A | 1.30 | 1.12 | UD | |
| E380A | 3.92 | |||
| D397A | 0.90 | 1.1 | UD | |
| D | ||||
| D17A | 5.22 | |||
| E40L | 4.05 | |||
| D88L | 1.87 | 2.3 | −25 | |
| E119A | 5.76 | |||
| D133A | 1.67 | 1.8 | −10 | |
| E153A | 4.46 | |||
| E | ||||
| E15A | 4.96 | |||
| E95A | 1.30 | 1.6 | UD | |
| E138A | 5.96 | |||
| E162G | 4.64 | |||
ΔΨ formation was measured in preparations of purified enzyme reconstituted into proteoliposomes, using Oxonol IV, as indicated in the Materials and Methods section. The spectrophotometric signal of Oxonol IV was calibrated by changing the diffusion potential of potassium. UD, undetectable (<−5mV).
Figure 2.
Formation of ΔΨ by wild-type Na+-NQR and acidic mutants reconstituted into proteoliposomes. A) Formation of ΔΨ by wild-type Na+-NQR showing the effects of CCCP (5µM), ETH-157 (5µM) and the omission of NaCl from the medium. B) Formation of ΔΨ by wild-type Na+-NQR and acidic mutants. The assay buffer contained: 100 µM Q-1, 100 mM HEPES, 100 mM NaCl (except where noted), 150 mM KCl, 1 mM EDTA, pH 7.5. The reaction was started by the addition of NADH (200 µM).
Kinetic properties of the wild- type Na+-NQR and acid mutants
In order to obtain a deeper insight into the effect of the mutations on the interaction of sodium with the enzyme, we studied the steady-state turnover at a range of sodium concentrations (Table 6). These experiments were performed with near-saturating concentrations of NADH (150 µM) and Q-1 (50 µM), in order to minimize rate limitation by substrates other than sodium. A Kmapp for sodium of about 1 mM was obtained for the wild-type enzyme, close to the value reported for other preparations (29). It is worth noting that the Kmapp for sodium obtained by steady state measurements is similar to the Kmapp for sodium of the rate limiting step in the reduction of Na+-NQR by NADH (29), indicating that this value could be close to the actual dissociation constant for sodium.
Table 6.
Apparent kinetic parameters of the wild-type Na+-NQR and acid mutants.
| Subunit | Mutant | Orientation | KmappNa+ (mM) | VmaxNa+/ VmaxnoNa+ |
|---|---|---|---|---|
| WT | 1.1 | 6.2 | ||
| B | E28A | P | 2.1 | 3.5 |
| E144L | N | 4.2 | 4.8 | |
| D346A | P | 0.9 | 1.4 | |
| D397A | N | >100 | - | |
| D | D88L | P | 1.4 | 3.1 |
| D133A | N | 9.3 | 2.8 | |
| E | E95A | N | 13.1 | 1.3 |
The Qred activity of the mutants was measured with different concentrations of NaCl using near saturating amounts of NADH (150 µM) and Q-1(50 µM). The kinetic parameters were calculated from the saturation curve using the Michaelis-Menten equation. The orientation was obtained from the topology studies published previously (15). N; negative side of the membrane (facing the cytosol), P; positive side of the membrane (facing the periplasmic space).
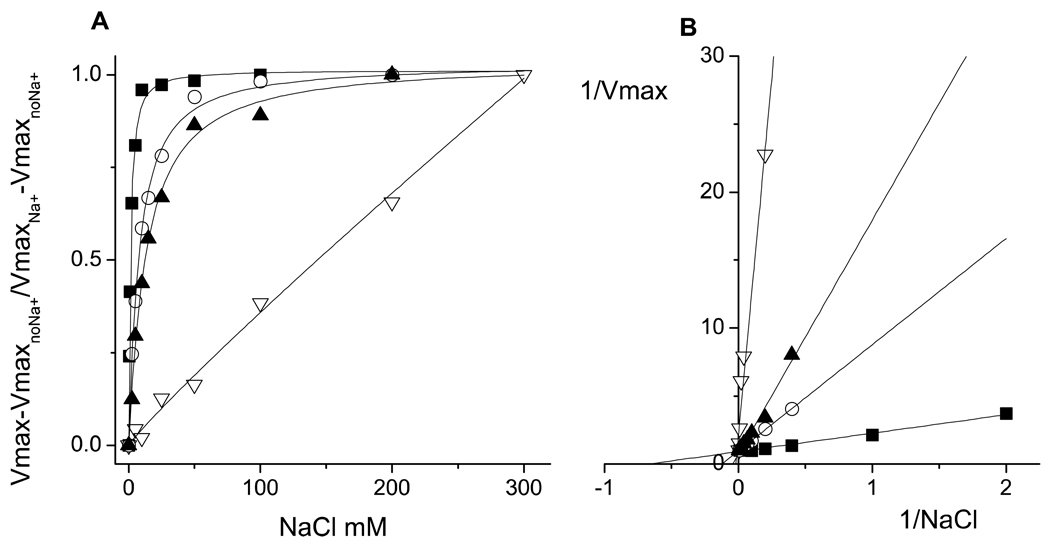
Three mutants, (NqrB-D397A, NqrD-D133A and NqrE-E95A), which are located in the part of the transmembrane helices facing the cytosol, showed an increase of 9 times or more in the Kmapp for sodium, suggesting that the mutations result in a change in the Na+ affinity of the enzyme (Table 6 and Figure 3). However, in the case of NqrB-D397A, the turnover rate increased linearly all the way to the highest concentration of sodium tested (300 mM). This could be consistent with either, 1. A very large change in the sodium affinity of the enzyme or, 2. Conditions in which sodium uptake is so severely restricted that the bimolecular process (collision of sodium with the enzyme) is always rate limiting. NqrB-E144L mutant could be considered as part of this group, because it produced a four-fold decrease in the Kmapp for sodium. However, the change is small compared with the other mutants, and in contrast with many of them, this mutation does not abolish the capacity for ion pumping (Table 5).
Figure 3.
A. Saturation kinetics for sodium of the wild-type (black squares) and the mutants NqrB-D397A (white triangles), NqrD-D133A (white circles) and NqrE-E95A (black triangles). For presentation purposes the VmaxnoNa+ was subtracted from the individual points of the saturation curves and then the data were normalized by the Vmax value obtained from the fit of the data of each mutant. In the case of the NqrB-D397A mutant data were normalized by the Vmax obtained with 300 mM NaCl, due to the apparently unsaturable behavior. B. Double reciprocal plots of the saturation kinetics. The analysis of the data was performed using non-linear curve fitting with a modified Michaelis-Menten equation (ν= Vmax•Na/(Kmapp + Na) + b) that contains the Vmax and Kmapp values, as well as the activity of the enzyme without sodium (b).
NqrB-E28A, NqrB-D346A and NqrD-D88L mutants caused little or no change in the Kmapp for sodium, in spite of the fact that the overall sensitivity of the turnover rate to sodium was significantly diminished as compared to the wild-type enzyme. This suggests that in these mutants, the sodium binding affinity of the enzyme has not changed, and that the mutations exert their effects at some other point in the reaction cycle. This would be consistent with a mechanism limited by restricted Na+ ejection, rather than uptake. It is interesting to note that all of the residues in the latter group are located near the positive side of the membrane facing the periplasm where sodium release takes place during turnover. Residues in the former group (NqrB-D397A, NqrD-D133A and NqrE-E95A) are all located near the negative side of the membrane, facing the cytoplasm, where sodium uptake takes place (Table 6). It is important to stress that the Km or the Kmapp of an enzyme does not directly reflect the dissociation constant for the substrate; because in both cases they are complex parameters that are composed of many rate constants, which vary depending on the mechanism of the enzyme. However, if we consider that these mutations had a selective effect and that they did not modify the binding of other substrates, or the stability of the protein, the Kmapp values could be used as a rough indication of the affinity for substrate.
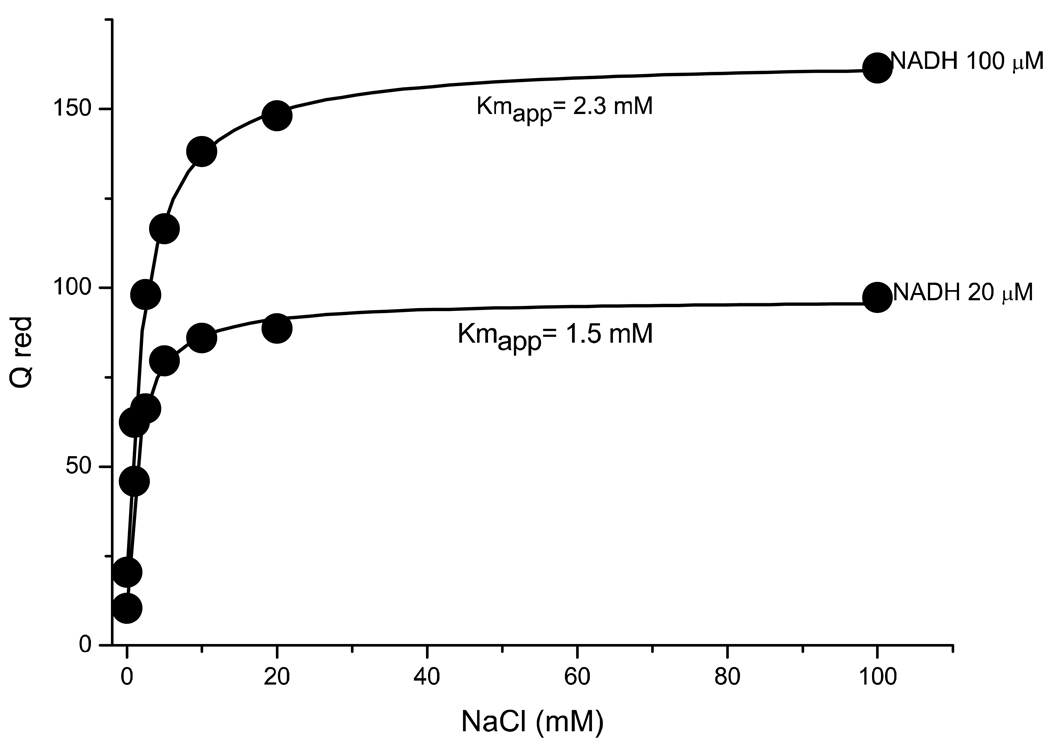
It was important to verify that the changes in Kmapp for sodium measured in these mutants did not arise as secondary effects of inhibition of the redox reaction in the mutants. To this end, we determined the Kmapp for sodium in the wild type enzyme at two different concentration of NADH (Figure 4). The Kmapp for sodium changes very little with changing NADH concentration, and in fact increases with increasing turnover rate, the opposite trend to that observed in the mutants. We also studied the effect of sodium concentration on the Km values for NADH and Q-1 for the wild type and selected mutants (NqrB-D346A and NqrB-D397A) enzyme. The Km values for NADH and Q-1 were essentially unaffected by sodium concentration, and were not significantly altered by the mutations (data not shown).
Figure 4.
Dependence of Qred activity of wild type Na+-NQR on sodium at two different concentrations of NADH.
DISCUSSION
In order to achieve sodium transport, Na+-NQR must be able to bind the cation with a physiologically relevant affinity and specificity, and sodium must be able to move across the membrane through a pore or a passageway. The gating or uni-directionality of sodium transport could be accomplished by coupling the energy produced in the redox reaction of the enzyme to structural changes in the protein, which would prevent ion back flow. The characterization of the sodium binding site (or sites) and sodium passageway is a prerequisite for understanding the coupling mechanism in Na+-NQR.
Negatively charged residues located in transmembrane segments of proteins are ideal candidates for the passageways or binding sites of cations in either pumps or ions channels. In fact, aspartates and glutamates are common components of sodium binding sites of both, soluble and membrane proteins (30).
Acid residues with a significant role in sodium translocation
Na+-NQR has a surprisingly large number of aspartates and glutamates in the membrane segments of subunits B, D and E. This indicates that, although these three subunits are very hydrophobic, they contain hydrophilic sites that could be important for sodium translocation. Our initial goal was to determine which of these amino acids have a significant role in enzyme activity and sodium binding.
There are 17 acid residues in transmembrane helices highly conserved across Na+-NQR sequences. Of these, 14 are conserved and semi-conserved in the related enzyme RNF, indicating that these residues are likely to be of special importance.
NqrB
Mutations of almost all the acid residues in the transmembrane helices of NqrB had a deleterious effect on sodium-dependent enzyme activity or sodium affinity, suggesting that these residues participate in binding or transporting sodium. Mutations of NqrB-D346 and NqrB-D397 both led to significant decreases in enzyme turnover and overall sodium sensitivity. However, in the case of NqrB-D346, which is located near to the periplasmic space, there was essentially no change in sodium affinity, while in the case of NqrB-D397, the sodium dependence of enzyme activity produced a straight line, (Figure 3) consistent with a very large decrease in the sodium affinity of the enzyme. Mutations of two other residues, NqrB-E380 and NqrB-E274, had only minimal effects on enzyme activity, suggesting that they do not play an important role in the mechanism of sodium pumping. These residues are both located near to the ends of transmembrane helices, where they are likely to interact with solvent in the periplasmic space. All of the other residues appear to be buried by at least one turn of the helix (3 residues) into the hydrophobic core of the membrane.
Two of these residues, which appear to have a significant role in enzyme activity, NqrB-E28 and NqrB-E144, are highly conserved in Na+-NQR but not in RNF. This lack of conservation probably reflects differences in the details of the sodium pumping mechanisms of the two complexes.
NqrD
Mutations of two of the six conserved acid residues in NqrD, E133 and D88, resulted in large changes in enzyme activity and sodium sensitivity. In the case of NqrD-D88, the Kmapp for sodium in the mutant is approximately the same as wild type, but in the case of NqrD-D133, the Kmapp for sodium is approximately 10 times higher than for wild-type enzyme. It is interesting to note that while NqrD-D88 appears to have an important role in enzyme function, it is predicted to be close to the end of a transmembrane helix, and likely to be in contact with solvent in the periplasmic space.
NqrE
Of the four conserved acid residues in NqrE, only NqrE-E95 appears to play a role in enzyme activity. Mutation of this residue, which is located in the hydrophobic part of the membrane, led to a significant decrease in enzyme turnover (Qred) and sodium sensitivity. Importantly, the apparent Km for sodium is about 10-fold higher than in the wild-type enzyme. Mutations of the three remaining acid residues produced very little effect on either enzyme turnover or sodium sensitivity. Two of these residues, NqrE-E138 and NqrE-E162 appear to be exposed to the cytoplasm, which may explain these results. However, NqrE-15 is located in the hydrophobic part of the membrane, and thus could have a role in stabilizing the structure.
A sodium binding site in Na+-NQR
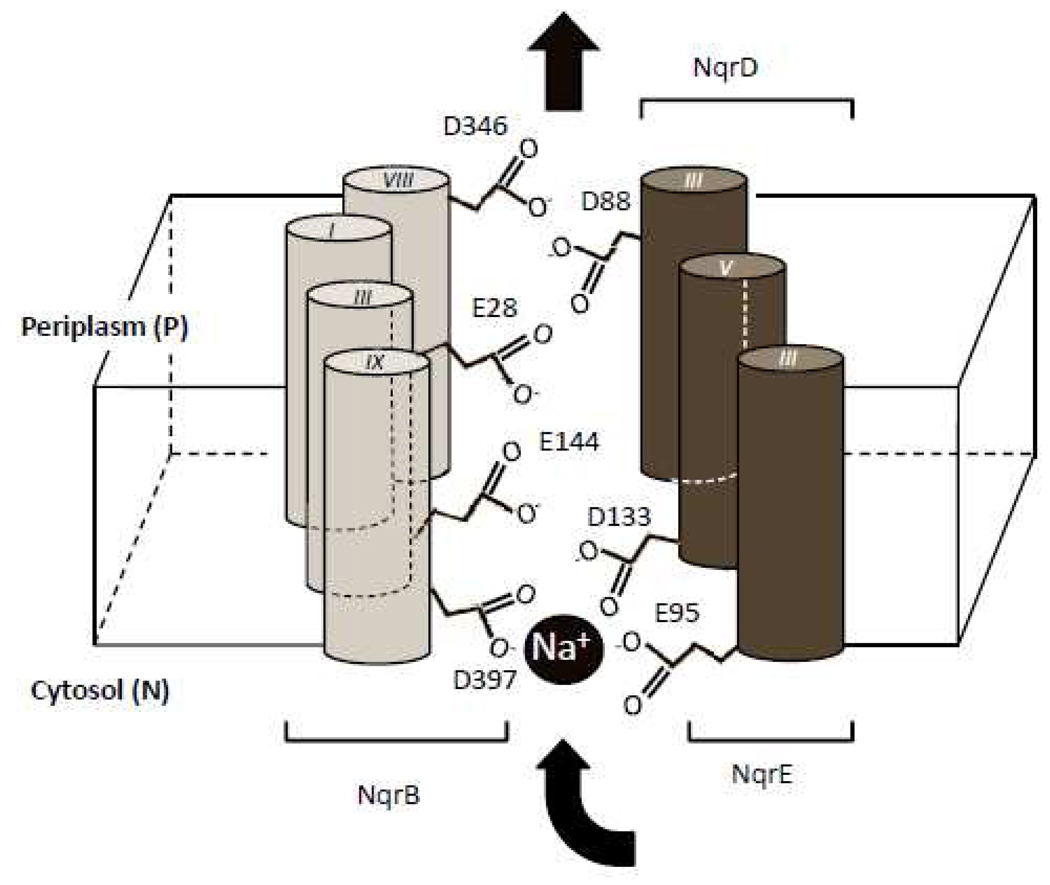
These results point to three residues (NqrB-D397, NqrD-D133 and NqrE-E95), all facing the cytoplasmic (negative) side of the membrane, which appear to serve an important role in the binding of sodium by Na+-NQR during turnover. Mutations of these three residues have large effects on the enzyme activity (Qred) and also significantly alter the apparent affinity of the enzyme for sodium (Figure 3). NqrB-D397 is perhaps the most important ligand in this putative Na+ binding site, since its mutant cannot be saturated with sodium under the experimental conditions tested, which is consistent with a very low affinity for the cation. NqrD-D133 and NqrE-E95 could participate in this putative binding site, since in both cases mutations increased Kmapp for sodium by approximately one order of magnitude (Table 6 and Figure 3).
NqrB-E144 may also have a role in sodium binding. Mutation of this residue (E144A) led to a four-fold increase in the Kmapp for sodium, (4.2 mM vs 1.1 mM for wild type). This change is smaller than for the other three residues suggesting a less important role as a ligand. Interestingly the value of Kmapp in this mutant is similar to that for wild-type V. harveyi Na+-NQR (3.3 mM) (29).
The side chains of NqrB-D397, NqrD-D133, NqrE-E95, and possibly NqrB-E144 may form a single sodium binding site. This would follow the pattern found in sodium binding sites I and II of Na+/K+-ATPase, which contain three and four acid residues, respectively (31). However, it has been shown that the presence of net negative charges is not absolutely necessary for sodium coordination; in fact sodium binding site I in the Leucine/Na+ co-transporter (30) and sodium binding site III of the Na+/K+ ATPase (31) contain only one net negative charge, while the second cation binding site of the Leucine/Na+ co-transporter does not include any negative charges (30). In these cases the other ligands are parts of serine or threonine residues, the protein backbone, and water molecules, carrying partial negative charges.
Nayal and Di Cera (22) studied ion binding sites in proteins and concluded that the main mechanism that determines the specificity of these sites is the number of ligands, the match between the size of the ion and the binding site, and the spatial distribution of the ligands. They determined that the optimal binding site for sodium consists of six ligands in an octahedral array (20). Thus, it is not surprising that the three main residues implicated in sodium binding in Na+-NQR are completely conserved in both Na+-NQR and RNF.
Our results also identified four additional residues (NqrB-E28, NqrB-E144, NqrB-D346 and NqrD-D88), mutants of which alter the enzyme activity (Qred) and its sodium sensitivity, but do not alter Kmapp for sodium. This suggests that these residues may be part of a structure that conducts sodium through the hydrophobic region of the membrane.
Figure 5 shows a possible arrangement of helices in which NqrB-D397, NqrD-D133 and NqrE-E95 take part in a sodium-binding site while NqrB-E144, NqrB-E28, NqrB-346 and NqrD-D88 participate in a structure that facilitates the passage of sodium through the hydrophobic membrane. The complete mechanism is likely to require additional residues and probably internal water. There are up to 15 conserved polar residues buried in the membrane; three are found in the NqrB subunit (S222, T345, N390), seven in NqrD (N18, N19, Q22, S55, S81, N111, C112) and five in NqrE (N16, C26, Q92, N120, C121). Conservation of these residues cannot be used to directly infer participation in sodium transport, since many of these amino acids play other roles in membrane proteins. For example, serine and threonine are common components of transmembrane helices, accounting for approximately 7% of all amino acid residues (17). These residues promote the formation of strong and specific associations between transmembrane helices, mediated by hydrogen bonds; some examples are found in halorhodopsin, Ca2+- ATPase and cytochrome c oxidase (17).
Figure 5.
Representation of a possible binding site and passageway of sodium in Na+-NQR.
All of the mutations studied in this paper involved replacement of acid residues by aliphatic ones, changing both the chemical properties and the size of the side-chain. At this point we cannot rule out the possibility that these mutations may affect the properties of the enzyme indirectly through structural perturbations. To investigate this possibility, we are now constructing new mutations at the most interesting sequence positions, with more conservative changes, for example D → E, D → N, etc. With these new mutations we hope to distinguish steric and chemical effects of the residues. However this initial survey has succeeded in identifying a subset of the acidic residues within transmembrane helices that are promising candidates for sodium binding sites in Na+-NQR. This represents an important initial step in the characterization of the coupling mechanism of Na+-NQR and also could be useful to understand the structural basis of the cation selectivity of the enzyme.
ACKNOWLEDGMENTS
We would like to thank Joel E. Morgan and Michael Shea for stimulating discussions and the critical reading of the manuscript.
Footnotes
This work is supported by National Institutes of Health Grant GM69936 (B.B).
ABBREVIATIONS
BCA, Bicinchonic acid; CCCP, carbonyl cyanide m-chlorophenylhydrazone; EDTA, ethylenediaminetetraacetic acid; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide; HEPES, (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid); LB, Luria-Bertani; Na+-NQR, Na+-pumping NADH:quinone oxidoreductase; RNF, Rhodobacter nitrogen fixing protein; Tris, tris(hydroxymethyl) aminomethane.
REFERENCES
- 1.Tokuda H, Udagawa T, Unemoto T. Generation of the electrochemical potential of Na+ by the Na+-motive NADH oxidase in inverted membrane vesicles of Vibrio alginolyticus. FEBS Lett. 1985;183:95–98. doi: 10.1016/0014-5793(85)80961-3. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi M, Nakayama Y, Unemoto T. Recent progress in the Na(+)-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. Biochim Biophys Acta. 2001;1505:37–44. doi: 10.1016/s0005-2728(00)00275-9. [DOI] [PubMed] [Google Scholar]
- 3.Bogachev AV, Verkhovsky MI. Na(+)-Translocating NADH:quinine oxidoreductase: progress achieved and prospects of investigations. Biochemistry (Mosc) 2005;70:143–149. doi: 10.1007/s10541-005-0093-4. [DOI] [PubMed] [Google Scholar]
- 4.Rich PR, Meunier B, Ward FB. Predicted structure and possible ionmotive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 1995;375:5–10. doi: 10.1016/0014-5793(95)01164-a. [DOI] [PubMed] [Google Scholar]
- 5.Turk K, Puhar A, Neese F, Bill E, Fritz G, Steuber J. NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae: functional role of the NqrF subunit. J Biol Chem. 2004;279:21349–21355. doi: 10.1074/jbc.M311692200. [DOI] [PubMed] [Google Scholar]
- 6.Barquera B, Hellwig P, Zhou W, Morgan JE, Hase CC, Gosink KK, Nilges M, Bruesehoff PJ, Roth A, Lancaster CR, Gennis RB. Purification and characterization of the recombinant Na(+)-translocating NADH:quinine oxidoreductase from Vibrio cholerae. Biochemistry. 2002;41:3781–3789. doi: 10.1021/bi011873o. [DOI] [PubMed] [Google Scholar]
- 7.Barquera B, Nilges MJ, Morgan JE, Ramirez-Silva L, Zhou W, Gennis RB. Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na(+)-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry. 2004;43:12322–12330. doi: 10.1021/bi048689y. [DOI] [PubMed] [Google Scholar]
- 8.Juárez O, Morgan JE, Barquera B. The electron transfer pathway of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J Biol Chem. 2009;284:8963–8972. doi: 10.1074/jbc.M809395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barquera B, Hase CC, Gennis RB. Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na(+) pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 2001;492:45–49. doi: 10.1016/s0014-5793(01)02224-4. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi M, Nakayama Y, Yasui M, Maeda M, Furuishi K, Unemoto T. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2001;488:5–8. doi: 10.1016/s0014-5793(00)02404-2. [DOI] [PubMed] [Google Scholar]
- 11.Nakayama Y, Yasui M, Sugahara K, Hayashi M, Unemoto T. Covalently bound flavin in the NqrB and NqrC subunits of Na(+)-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2000;474:165–168. doi: 10.1016/s0014-5793(00)01595-7. [DOI] [PubMed] [Google Scholar]
- 12.Barquera B, Ramirez-Silva L, Morgan JE, Nilges MJ. A new flavin radical signal in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae: An EPR/ENDOR investigation of the role of the covalently bound flavins in subunits B and C. Journal of Biological Chemistry. 2006;281:36482–36491. doi: 10.1074/jbc.M605765200. [DOI] [PubMed] [Google Scholar]
- 13.Barquera B, Zhou W, Morgan JE, Gennis RB. Riboflavin is a component of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. 2002;99:10322–10324. doi: 10.1073/pnas.162361299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juarez O, Nilges MJ, Gillespie P, Cotton J, Barquera B. Riboflavin is an active redox cofactor in the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. Journal of Biological Chemistry. 2008;283:33162–33167. doi: 10.1074/jbc.M806913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffy EB, Barquera B. Membrane topology mapping of the Na+-pumping NADH: quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J Bacteriol. 2006;188:8343–83451. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biegel E, Schmidt S, Muller V. Genetic, immunological and biochemical evidence for a Rnf complex in the acetogen Acetobacterium woodii. Environ Microbiol. 2009;11:1438–1443. doi: 10.1111/j.1462-2920.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- 17.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 18.Facciotti MT, Rouhani-Manshadi S, Glaeser RM. Energy transduction in transmembrane ion pumps. Trends Biochem Sci. 2004;29:445–451. doi: 10.1016/j.tibs.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Page MJ, Di Cera E. Role of Na+ and K+ in enzyme function. Physiol Rev. 2006;86:1049–1092. doi: 10.1152/physrev.00008.2006. [DOI] [PubMed] [Google Scholar]
- 20.Nayal M, Di Cera E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J Mol Biol. 1996;256:228–234. doi: 10.1006/jmbi.1996.0081. [DOI] [PubMed] [Google Scholar]
- 21.Rigaud JL, Pitard B, Levy D. Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochem Biophys Acta. 1995;1231:223–246. doi: 10.1016/0005-2728(95)00091-v. [DOI] [PubMed] [Google Scholar]
- 22.Verkhovskaya ML, Garcìa-Horsman A, Puustinen A, Rigaud JL, Morgan JE, Verkhovsky MI, Wikström M. Glutamic acid 286 in subunit I of cytochrome bo3 is involved in proton translocation. Proc Natl Acad Sci U SA. 1997;94:10128–10131. doi: 10.1073/pnas.94.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apell HJ, Bersch B. Oxonol VI as an optical indicator for membrane potentials in lipid vesicles. Biochim Biophys Acta. 1987;903:480–494. doi: 10.1016/0005-2736(87)90055-1. [DOI] [PubMed] [Google Scholar]
- 24.Smith JC. Potential-sensitive molecular probes in membranes of bioenergetic relevance. Biochim Biophys Acta. 1990;1016:1–28. doi: 10.1016/0005-2728(90)90002-l. [DOI] [PubMed] [Google Scholar]
- 25.Imkamp F, Biegel E, Jayamani E, Buckel W, Muller V. Dissection of the caffeate respiratory chain in the acetogen Acetobacterium woodii: identification of an Rnf-type NADH dehydrogenase as a potential coupling site. J Bacteriol. 2007;189:8145–8153. doi: 10.1128/JB.01017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi M, Unemoto T. Characterization of the sodium-dependent respiratory chain NADH: quinone oxidoreductase of the marine bacterium, Vibrio alginolyticus, in relation to the primary sodium pump. Biochim. Biophys. Acta. 1984;767:470–477. [Google Scholar]
- 27.Pfenninger-Li XD, A, SPJ, van Belzen R, Dimroth P. NADH:ubiquinone oxidoreductase of Vibrio alginolyticus: purification, properties, and reconstitution of the Na+ pump. Biochemistry. 1996;35:6233–6242. doi: 10.1021/bi953032l. [DOI] [PubMed] [Google Scholar]
- 28.Steuber J, Krebs W, Dimroth P. The Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio alginolyticus--redox states of the FAD prosthetic group and mechanism of Ag+ inhibition. Eur J Biochem. 1997;249:770–776. doi: 10.1111/j.1432-1033.1997.t01-2-00770.x. [DOI] [PubMed] [Google Scholar]
- 29.Bogachev AV, Bertsova YV, Barquera B, Verkhovsky MI. Sodium-dependent steps in the redox reactions of the Na+-motive NADH:quinone oxidoreductase from Vibrio harveyi. Biochemistry. 2001;40:7318–7323. doi: 10.1021/bi002545b. [DOI] [PubMed] [Google Scholar]
- 30.Gouaux E, Mackinnon R. Principles of selective ion transport in channels and pumps. Science. 2005;310:1461–1465. doi: 10.1126/science.1113666. [DOI] [PubMed] [Google Scholar]
- 31.Rakowski RF, Sagar S. Found: Na(+) and K(+) binding sites of the sodium pump. News Physiol Sci. 2003;18:164–168. doi: 10.1152/nips.01441.2003. [DOI] [PubMed] [Google Scholar]