Abstract
Fast-transmitting vertebrate axons are electrically insulated with multiple layers of nonconductive plasma membrane of glial cell origin, termed myelin. The myelin membrane is dominated by lipids, and its protein composition has historically been viewed to be of very low complexity. In this review, we discuss an updated reference compendium of 342 proteins associated with central nervous system myelin that represents a valuable resource for analyzing myelin biogenesis and white matter homeostasis. Cataloging the myelin proteome has been made possible by technical advances in the separation and mass spectrometric detection of proteins, also referred to as proteomics. This led to the identification of a large number of novel myelin-associated proteins, many of which represent low abundant components involved in catalytic activities, the cytoskeleton, vesicular trafficking, or cell adhesion. By mass spectrometry-based quantification, proteolipid protein and myelin basic protein constitute 17% and 8% of total myelin protein, respectively, suggesting that their abundance was previously overestimated. As the biochemical profile of myelin-associated proteins is highly reproducible, differential proteome analyses can be applied to material isolated from patients or animal models of myelin-related diseases such as multiple sclerosis and leukodystrophies.
Keyword: Oligodendrocyte, Leukodystrophy, Myelin, Internode, Proteome, Proteomics, Cytoskeleton, Neurodegeneration, Proteolipid protein, Myelin basic protein
Introduction
Neuronal signal propagation in vertebrates is sped up by the electrical insulation of axons with an ensheathing, specialized glial plasma membrane: myelin. Myelination of axons reduces their transverse capacitance and increases their transverse resistance [1]. Insulation is achieved by the multilayered arrangement of the myelin membrane (Fig. 1) and its special molecular composition, mainly its very high lipid content. In myelinated axons, action potentials are restricted to periodically spaced small segments spared from coverage with myelin, termed the nodes of Ranvier [2]. In the central nervous system (CNS), any individual oligodendrocyte myelinates up to 50 axon segments, termed internodes [3]. Oligodendrocyte precursor cell division, migration, and regular alignment along the axons have been recently visualized in vivo in zebrafish [4], which today complement rodents as an important model organism for myelin research [5–8]. Myelin formation proceeds with outgrowth and retraction of glial cell processes, target axon recognition, stabilization of cellular contacts, rapid biosynthesis and trafficking of lipid and protein constituents of the myelin membrane, and its organization as a multilayered structure around the axon [9, 10]. Once myelinated, axons become dependent on glial support [11]. Some of the molecules involved in myelin development and function are known but a detailed molecular picture has not been gained yet.
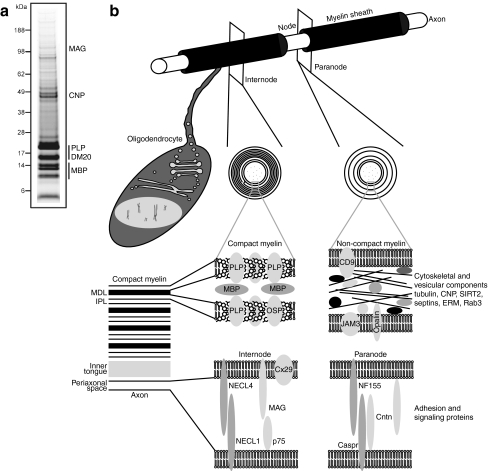
Fig. 1.
CNS myelin. a Purified mouse brain myelin was one-dimensionally separated in a 4–12% Bis–Tris gradient gel using a morpholineethanesulfonic acid buffer system. Proteins were visualized by colloidal Coomassie staining. Bands constituted by abundant myelin proteins are annotated. b Schematic depiction of an oligodendrocyte myelinating an axon, cross-sections in the internodal and paranodal segments, and subcellular localization of myelin proteins. Structural proteins of compact myelin (middle), cytoskeletal and vesicular proteins located in uncompacted regions (right), and adhesion proteins mediating association with the axon (bottom) are shown. CNP 2′,3′-cyclic nucleotide phosphodiesterase, Cntn contactin, Caspr contactin-associated protein, Cx29 connexin 29 kDa, DM20 small splice isoform of PLP, ERM ezrin, radixin, moesin, IPL intraperiod line, JAM3 junctional adhesion molecule 3, MAG myelin-associated glycoprotein, MBP myelin basic protein, MDL major dense line, Necl nectin-like protein, NF155 neurofascin 155 kDa, OSP oligodendrocyte-specific protein/claudin-11, PLP proteolipid protein, Rab3 Ras-related protein Rab3, SIRT2 sirtuin 2
That CNS myelin is important for normal sensation, cognition, and motor function is obvious considering that myelin-related disorders often affect humans lethally. Besides the inflammatory demyelinating disease multiple sclerosis [12], there are genetically inherited disorders that affect CNS myelin, collectively termed leukodystrophies [13]. This heterogeneous group of diseases is characterized by the loss of motoric, sensory, and mental capabilities and the susceptibility to seizures. A detailed knowledge of the molecular expression profiles of oligodendrocytes and myelin will be crucial to understand the pathomechanisms of white matter diseases. For example, the mRNAs [14–16] and proteins expressed in cultivated oligodendrocytes [17] and oligodendroglial exosomes [18] have been recently examined. This review focuses on systematic analyses of the molecular composition of mammalian CNS myelin, while no such compendium of peripheral nervous system (PNS) myelin proteins has been published yet. Proteomics approaches to myelin provide a valuable resource to understand its biogenesis, function, and pathology. Although only a few comparative studies have been reported to date, novel insights into the molecular basis of myelin-related diseases are beginning to emerge.
A Myelin-Enriched Fraction from the Central Nervous System
A comparatively simple method is available for the isolation of a myelin-enriched fraction from the CNS. Biochemically, myelin is defined as the lightweight membranous material accumulating at the interface between 0.32 and 0.85 M sucrose after sequential ultracentrifugation combined with osmotic shocks [19, 20]. The most commonly used protocol starts from brain homogenate contained in 0.32 M sucrose as the top layer, “spinning-down” myelin to accumulate at the interface with the bottom 0.85 M sucrose layer. One valuable modification is “floating-up” of myelin starting from brain homogenate contained in a more concentrated sucrose solution as the bottom layer (0.85, 1.2, 1.44, or 2 M). During ultracentrifugation, myelin also accumulates at the interface between the upper 0.85 and 0.32 M sucrose layers, while other fractions of interest assemble at higher sucrose concentrations. This method allows the simultaneous isolation of other brain fractions such as rough microsomes [21] or axogliosomes [22, 23]. The lightweight fraction from the interphase between 0.32 and 0.85 M sucrose is the most frequently used one for biochemical and proteomic experiments. This fraction is enriched in the most abundant proteins of compact myelin, proteolipid protein (PLP), and myelin basic protein (MBP), and as revealed by electron microscopy, mainly contains multilamellar membranes with a periodicity comparable to that of myelin in native or perfused brains [24, 25]. However, we suggest to term this fraction “myelin-enriched” rather than “compact myelin”, as it also contains proteins from the noncompacted cytosolic channels in myelin (i.e., adaxonal and paranodal myelin) and proteins associated with the axonal membrane. Myelin purification is very reproducible across different laboratories, even when applied to different species (e.g., mouse–rat) or to mutant mice with altered myelin protein or lipid composition, such as CnpCre/+*Fdftflox/flox [26], Ugt3a1null [27], Arsanull [28], and Plpnull [29] (see below). Thus, the method has proven to be very robust, explaining why the original protocol from the early 1970s is still in common use. It is generally assumed that myelin purification relies on its special lipid content and composition.
Myelin Lipids
The molecular composition of myelin differs from other plasma membranes in that it contains 70–75% of its dry weight as lipid, unusually high compared to other eukaryotic plasma membranes. Also, its molar ratio of lipids with approximately 2:2:1:1 for cholesterol/phospholipid/galactolipid/plasmalogen [30, 31] distinguishes myelin from other cellular membranes. The abundance of cholesterol within a membrane affects its biophysical properties, including fluidity and curving [32]. Cholesterol has earlier been identified as unusually enriched in myelin and constitutes 24–28% of the total myelin lipids [19]. That the cellular cholesterol supply is rate-limiting for myelin membrane biogenesis has been shown in mice lacking squalene synthase (also termed farnesyl diphosphate farnesyl transferase [FDFT]) exclusively in myelinating glia [26]. FDFT mediates a crucial step of cholesterol biosynthesis. CNS myelination is severely delayed in CnpCre/+*Fdft1flox/flox mice, and that any myelin made in these mice is likely due to compensatory cholesterol uptake from other cells [26].
The biophysical properties of myelin are also influenced by its unusually high concentration of the galactolipids galactosylceramide (GalC), its sulfated form 3-O-sulfogalactosylceramide (SGalC), and their hydroxylated forms GalC-OH and SGalC-OH. Together, they add up to 20–26% of total myelin lipids. Myelination is moderately delayed in mice lacking UDP-galactose:ceramide galactosyltransferase (Ugt3a1), an enzyme required for galactolipid synthesis. Additionally, impaired glia–axonal interactions at the paranodes were observed [27, 33, 34]. Paranodal disruption was at least partly due to the lack of SGalC and hydroxylated galactolipids, since the long-term integrity of the sodium channel domain of the nodes of Ranvier was also impaired in mice lacking galactosylceramide-3-O-sulfotransferase (Gal3st1), the enzyme converting GalC into SGalC [35–37], and late onset myelin degeneration was also reported for mice lacking fatty acid 2-hydroxylase (Fa2h), the enzyme hydroxylating GalC and SGalC [38]. Absence of functional arylsulfatase A (ARSA), the enzyme degrading SGalC, causes metachromatic leukodystrophy (MLD), illustrating that a regulated galactolipid metabolism is required for long-term integrity of the white matter. SGalC accumulation and many pathological features of MLD are modeled in Arsanull mice and in transgenic mice overexpressing Ugt3a1 or Gal3st1 in neurons or oligodendrocytes [28, 39, 40]. Sulfatide metabolism with respect to myelin and MLD pathology was recently reviewed [41].
Also, the plasmalogen class of phospholipids is associated with white matter disease. Plasmalogens are ether-linked (as opposed to ester-linked) phospholipids, the main species being ethanolamine–plasmalogen. They are ubiquitous structural components of mammalian cell membranes and amount to 12–15% of total myelin lipid [19] and, when processed by plasmalogen-selective phospholipase A2, give rise to the second messengers arachidonic acid and eicosanoids [42]. At low concentrations, these metabolites have trophic effects, but at high levels, they are cytotoxic and may induce inflammation [43]. The reactivity of the alkenyl ether bond makes plasmalogens more susceptible to oxidative reactions than their fatty acid ester analogs. Thus, myelin plasmalogens may act as endogenous antioxidants protecting cells from oxidative stress [44]. Disrupted activity of peroxisomal plasmalogen synthesizing enzymes results in peroxisomal biogenesis disorders such as rhizomelic chondrodysplasia punctata (RCDP) in which hypomyelination of the optic nerve has been observed. Decreased plasmalogen levels [45, 46] and increased levels of reactive oxygen species [47, 48] may also contribute to the demyelination in X-linked adrenoleukodystrophy caused by the mutated peroxisomal transporter ABCD1, suggesting that a normal plasmalogen metabolism may prevent peroxisomal- and myelin-related disease. Mice lacking dihydroxyacetonephosphate acyltransferase (DAPAT) model several aspects of the RCDP pathology, including optic nerve hypoplasia [49]. Interestingly, the association of flotillin-1 and contactin with plasmalogen-deficient brain membrane microdomains was diminished in DAPATnull mice [49], suggesting that the local concentration of membrane lipids dictates the association of particular proteins.
Association of Myelin Lipids and Proteins
Cholesterol assembles with galactolipids and plasmalogens within the plane of the membrane, but how they are enriched to the levels found in myelin is unknown. It has been suggested that lipids are targeted to future myelin membrane by their association with myelin-bound proteins [9]. SGalC appears to be an example to the contrary. SGalC is associated with myelin and lymphocyte protein (MAL) [50]. Lack of SGalC and lack of MAL lead to similar paranodal malformation [35, 51]. The subcellular trafficking of MAL, as well as its abundance in myelin, is determined by SgalC [28], whereas SGalC abundance is not altered in Malnull myelin [51]. It is likely that other myelin proteins are also incorporated into the sheath by attachment with future myelin membrane because of its special lipid composition. Thus, whether myelin proteins dictate the fate of lipids or vice versa may not be generalized. It appears likely that the association of both molecule classes results in each other’s control of abundance and trafficking.
That myelin lipids and proteins are closely associated was suggested earlier after the characterization of two types of protein fractions isolated from the white matter based on their resistance to aqueous or organic solvents or to enzymatic proteolysis. One fraction behaved as a lipid with regard to its solubility and was termed PLP [52, 53]. PLP was later identified to be the most abundant protein of mammalian CNS myelin. It has a high affinity to phospholipids and cholesterol [54–56], and impaired interactions of mutant PLP with membrane lipids are a likely key step in the molecular pathogenesis of the leukodystrophy Pelizaeus–Merzbacher disease [57]. The other fraction, termed trypsin-resistant protein residue, was insoluble in organic solvent and attached to the membrane lipid phosphatidylinositol phosphate [58, 59]. The application of extraction methods by Folch became commonly used to categorize myelin proteins according to their biophysical properties.
More recently, the myelin-enriched brain fraction has been chemically subfractionated by differential detergent extraction at low temperatures, resulting in distinct nonidentical but overlapping assemblies of myelin-associated proteins and lipids that were suggested to represent myelin subcompartments [60, 61]. Cholesterol- and galactolipid-rich membrane microdomains (also referred to as “lipid rafts”) have been suggested to deliver myelin proteins to the plasma membrane [62–64]. The relevance of applying the analysis of biochemical characteristics established for membrane microdomains to such a large structure as myelin has remained debated. However, it is widely accepted now that lipid-associated cell signaling molecules, such as the protein tyrosine kinase fyn, have central roles in myelination [65, 66].
In oligodendroglial processes, fyn is activated by axonal signals via integrin alpha6beta1 [67]. Among other fyn substrates [68, 69], the protein translation repressor heterogeneous nuclear ribonucleoprotein (hnRNP) A2 upon phosphorylation is released from its binding site in the 3′UTR of mRNA encoding MBP [70], the second-most abundant myelin protein. hnRNP A2 binding represses translation during the translocation of MBP mRNA to distal sites of the cell [71] where newly translated MBP is directly incorporated into the extending oligodendroglial process [21, 72]. It is generally assumed that MBP mediates the adhesion of the cytoplasmic surfaces between the individual layers of compact myelin [73] via binding of its many basic residues with the negatively charged headgroups of membrane lipids. Indeed, membrane association of MBP is controlled by the membrane lipid phosphatidylinositol-(4,5)-bisphosphate [74–76]. For over 30 years, it has been known that MBP is highly heterogeneous due to alternative splicing and multiple post-translational modifications (PTMs) [77]. More recently, modern mass spectrometric techniques have been used to compare the PTMs of MBP from normal and multiple sclerosis brains with respect to methylation, phosphorylation, and arginine deimination [78]. PTM alterations affect charge, conformation, and hydrogen bonding of MBP, which may modulate its affinity to the myelin membrane and play a role in myelin compaction and in the pathogenesis of demyelinating diseases. MBP is the only myelin protein that has been shown to be essential for myelin formation, as became obvious with the analysis of the natural mouse mutant shiverer and the rat mutant long evans shaker [79, 80], which are severely hypomyelinated. Interestingly, mice lacking fyn are also hypomyelinated [81, 82], likely due to affected translational regulation of MBP expression [70, 83]. Together, a multitude of factors affects mRNA transcription and transport, translation at axonal contact sites, or membrane binding of MBP, and we speculate that several myelin proteins with yet unidentified roles affect MBP abundance and function.
Systematic Analysis of the CNS Myelin Protein Composition
The relative abundance of myelin proteins has previously been calculated based on their binding to Buffalo black [84], Fast green [85], or Coomassie blue [86] after separation in one-dimensional (1D) sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE). In these measurements, a small number of proteins was determined to be extraordinarily abundant in CNS myelin. PLP and its smaller splice isoform DM20 accounted for 30–45% of total myelin protein, two of the four MBP splice isoforms for 22–35%, 2′,3′-cyclic nucleotide 3′-phosphodiesterase (CNP) for 4–15%, and all remaining proteins for 5–25% [19, 85, 87, 88]. Similarly, PNS myelin is also dominated by two proteins, myelin protein zero (MPZ, P0) and MBP, which have been estimated to account for 50–70% and 15%, respectively [89]. In comparison, the most abundant proteins in a brain fraction enriched for synaptic vesicles are synaptobrevin 2 and synaptophysin, which constitute 8% and 10% of the total synaptic vesicle proteins, respectively, as revealed by quantitative immunoblotting [90]. How and why myelin proteins are enriched to their unusual relative abundance is unclear, considering that PLP and CNP are not essential for the formation of normal amounts of CNS myelin [29, 91, 92].
Various proteomic techniques have been applied towards the systematic protein composition analysis of the myelin-enriched fraction. Traditionally, first insights into proteomes of subcellular structures often come from two-dimensional (2D) protein maps generated by utilizing isoelectric focusing (IEF) with immobilized pH gradients in the first and SDS-PAGE in the second dimension (2D-IEF/SDS-PAGE) (Fig. 2a). Proteins of interest are then excised from the gel, proteolytically digested in situ, and finally, identified by mass spectrometry (MS) [93]. Due to its high resolving power, 2D-IEF/SDS-PAGE can be routinely applied for profiling of proteins from complex mixtures and, as protein integrity is retained, also leads to information on protein abundance and processing [94]. However, major shortcomings of 2D-IEF/SDS-PAGE concern a limited dynamic range, the display of basic and hydrophobic proteins, and—most importantly—the under-representation of membrane proteins. As myelin is dominated by MBP (a highly basic protein) and PLP (a hydrophobic tetraspan protein), incremental improvements in 2D-IEF/SDS-PAGE technology were required before the first 2D mapping of myelin was presented [95]. By using the zwitterionic detergent amidosulfobetaine-14 (ASB-14) instead of the most commonly used 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) [96], it was possible to solubilize myelin proteins much more effectively and to identify 98 proteins (91 by MS and seven by immunoblotting) in the myelin-enriched fraction from mouse CNS [95]. This crucial effect of the solubilization conditions is further underscored by two more recent 2D-IEF/SDS-PAGE mapping studies of similar input material. Thirty-eight myelin-associated proteins were identified in one study after CHAPS solubilization [97], but 131 proteins were identified in another study with ASB-14 [25]. Thus, at least in the presence of appropriate detergents, myelin can now be considered as well accessible by 2D-IEF/SDS-PAGE, which not only facilitates protein cataloging but also paves the way for differential myelin proteomics on the basis of the 2D differential fluorescence intensity gel electrophoresis technology (2D-DIGE, see below). It is important to note that all conventional 2D mapping approaches mentioned above failed to appropriately display relatively abundant transmembrane myelin marker proteins such as PLP, myelin-associated glycoprotein (MAG) [98], myelin oligodendrocyte glycoprotein (MOG) [99], tetraspanin 2 [100], M6B [101], or oligodendrocyte-specific protein (OSP/claudin-11) [102–104]. A potential remedy is to perform the first dimension separation as nonequilibrium pH gradient electrophoresis for the 2D mapping of myelin proteins [105]. However, although this method appeared promising particularly for displaying highly basic proteins, it did not get as popular as 2D-IEF/SDS-PAGE with immobilized pH gradients, mainly due to limitations in reproducibility and resolution.
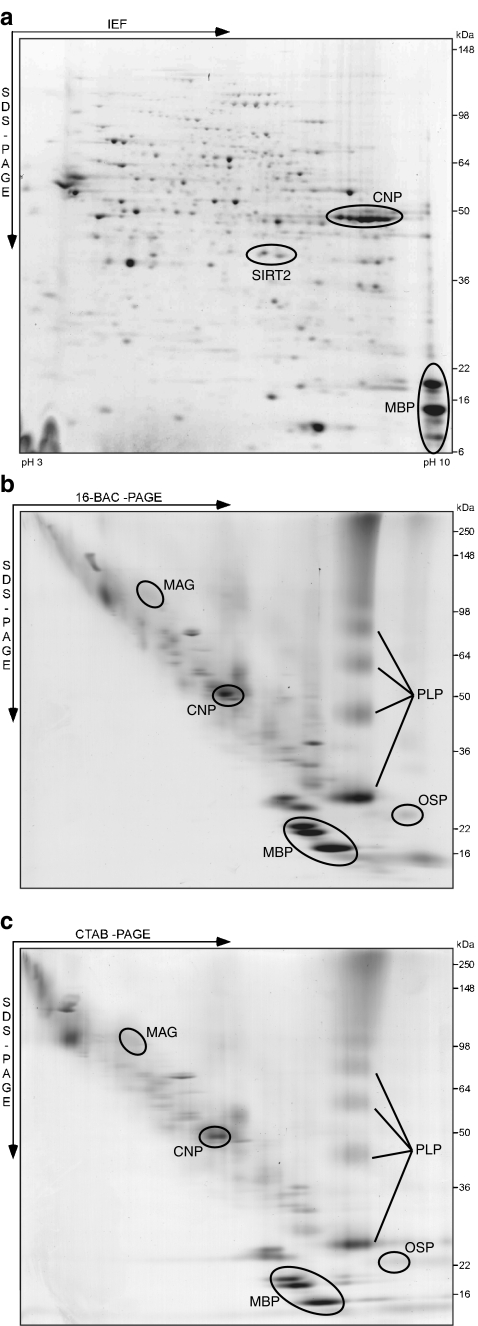
Fig. 2.
Gel-based myelin proteome maps. Purified mouse brain myelin was two-dimensionally separated in different gel systems. Proteins were visualized by colloidal Coomassie staining, and spots constituted by selected myelin proteins are indicated. a 2D-IEF/SDS-PAGE with IEF in a nonlinear pH gradient (pH 3–10) as the first and gradient SDS-PAGE (8–16% acrylamide) as the second dimension. To improve resolution, myelin was delipidated and precipitated by a methanol/chloroform treatment prior to IEF [25]. b 2D-16-BAC/SDS-PAGE with separation in a 16-BAC gel (10% acrylamide) as the first and gradient SDS-PAGE (8–16% acrylamide) as the second dimension. c 2D-CTAB/SDS-PAGE with separation in a CTAB gel (10% acrylamide) as the first and gradient SDS-PAGE (8–16% acrylamide) as the second dimension. To deplete soluble and membrane-associated proteins, myelin was subjected to a multistep wash procedure before separation [25]. 16-BAC and CTAB resulted in similar spot patterns. 2D-IEF/SDS-PAGE provides good resolution but basic, hydrophobic, and transmembrane proteins are under-represented. 2D-16-BAC/SDS-PAGE and 2D-CTAB/SDS-PAGE lead to efficient representation of basic, hydrophobic, and transmembrane proteins but have a lower resolution since separation occurs by protein size in both dimensions
More complete proteome coverage while retaining the benefits of displaying intact proteins can be reached by the additional use of alternative 2D gel systems. Here, the charge-dependent separation in the first dimension (i.e., the IEF) is replaced by a size-dependent separation in the presence of cationic detergents such as 16-benzyldimethyl-n-hexadecylammonium chloride (16-BAC; Fig. 2b) [106] or cetyltrimethylammonium bromide (CTAB; Fig. 2c) [107]. Due to the similar separation principle in both dimensions, proteins are typically dispersed along a diagonal rather than distributed over the entire gel area. Accordingly, these gel systems have a lower resolution compared to 2D-IEF/SDS-PAGE, but can resolve highly basic and even membrane-spanning proteins [108]. Application of 2D-16-BAC/SDS-PAGE to mouse CNS myelin resulted in the identification of 62 proteins and readily enabled displaying of the transmembrane myelin proteins PLP, MAG, MOG, and OSP/claudin-11 [25]. Thus, the combination of 2D-IEF/SDS-PAGE and 2D-16-BAC/SDS-PAGE has, so far, yielded the most comprehensive gel-based proteome compendium of mouse CNS myelin, consisting of 162 nonredundant proteins [25]. Further technical refinements of the method were established in a recent systematic evaluation of five different cationic detergents for the 2D gel electrophoresis of myelin proteins. Here, 16-BAC was the most effective agent for the separation of myelin proteins in the first dimension, while CTAB was most effective for their solubilization [109, 110]. As resolution improves, 2D gel electrophoresis with cationic detergents may be combined with the DIGE technology as a future tool for monitoring abundance changes of highly basic and membrane-spanning myelin proteins [111].
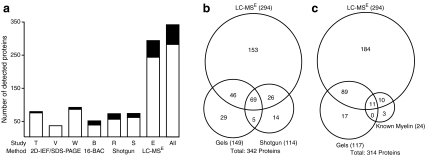
To overcome the limitations of gel-based proteomic methods, in particular those of 2D-IEF/SDS-PAGE, gel-free techniques, commonly referred to as shotgun approaches, have emerged in recent years [93, 112]. Here, separation at the level of intact proteins is omitted and the protein preparation is proteolytically digested at the expense of information related to protein integrity, such as protein size and charge. Separation takes place at the level of proteolytic peptides before interfacing with MS. The tremendous complexity of such peptide mixtures requires a high resolving power and is, therefore, often addressed by the application of 2D liquid chromatography (2D-LC), usually consisting of strong cation exchange in the first and reversed-phase chromatography in the second dimension. In the first application of shotgun proteomics to the myelin-enriched fraction from the mouse CNS [97], 93 proteins were identified resulting—by combination with 2D-IEF/SDS-PAGE (see above)—in a myelin proteome compendium consisting of 103 proteins. The application of a similar shotgun approach to a myelin-enriched fraction from rat CNS led to the identification of 97 myelin proteins [23]. Both shotgun approaches yielded quite a high overlap of approximately 50% with the so far most comprehensive gel-based library [25] and contained transmembrane myelin proteins such as PLP, MAG, and MOG.
Relative Abundance of Myelin Proteins
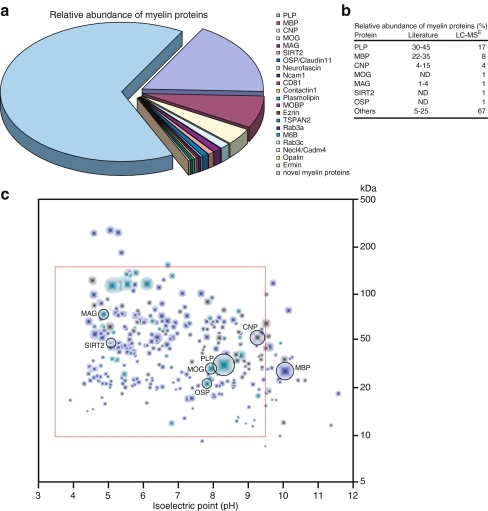
To understand myelin biogenesis and pathology, a comprehensive knowledge of the proteins associated with myelin is a prerequisite. We have confirmed and expanded the previous myelin protein compendia by applying nanoscale 1D ultra performance liquid chromatography (1D-UP-LC) separation coupled to detection with a quadrupole time-of-flight (QTOF) mass spectrometer (Tenzer et al., unpublished). Data were acquired by LC-MS using an alternating low (MS) and elevated (MSE) collision energy mode of acquisition (LC-MSE), which allows simultaneous identification and label-free relative quantification of the proteins in the sample [113–115]. The identified peptides were annotated to a total of 294 myelin-associated proteins (Table 1) based on a minimum of two peptides per protein with an effective false-positive rate of <0.2%. They showed a very good overlap of 141 proteins that were also detected in previous myelin proteome analyses and included several established myelin markers (Table 1 and Fig. 3). We have calculated the relative abundance of the myelin-associated proteins based on the average intensity of the three most abundant peptides per protein. In the few cases where only two peptides were identified, their average intensity was used. Strikingly, PLP, MBP, and CNP constituted only 17%, 8%, and 4% of the total myelin-associated proteins, respectively (Fig. 4). All previously known myelin proteins together constituted 35%, while newly identified myelin-associated proteins accounted for 65%. These quantifications take into question previous estimates based on conventional techniques (Fig. 4b and see above). We suggest that the complexity of myelin protein composition has been overlooked because low abundant proteins did not constitute significant bands on gels when compared to the highly abundant PLP and MBP due to limitations concerning gel separation and/or protein staining.
Table 1.
The CNS myelin proteome
| Protein name | ID | Gene | Reference |
|---|---|---|---|
| A: Known myelin proteins | |||
| CD81 | P35762 | Cd81 | E |
| CD9 | P40240 | Cd9 | ND |
| Claudin 11, OSP | Q60771 | Cldn11 | B,S,T,E |
| CNP | P16330 | Cnp | W,B,S,R,T,E |
| Contactin 1 | P12960 | Cntn1 | B,S,R,T,E |
| Ermin | Q5EBJ4 | Ermn | E |
| Ezrin | P26040 | Ezr | W,T,E |
| Glycoprotein M6B | P35803 | Gpm6b | E |
| Myelin and lymphocyte protein | O09198 | Mal | ND, T (blot) |
| Myelin-associated glycoprotein | P20917 | Mag | B,S,R,E |
| Myelin basic protein | P04370 | Mbp | W,B,S,V,R,E |
| Myelin oligodendrocyte glycoprotein | Q61885 | Mog | B,S,R,E |
| Myelin protein zero, P0 | P27573 | Mpz | R |
| Myelin proteolipid protein | P60202 | Plp1 | B,S,R,T,E |
| Myelin/oligodendrocyte basic protein | Q9D2P8 | Mobp | E |
| Necl1, Ig superfamily member 4b | Q99N28 | Cadm3 | S |
| Necl4, Ig superfamily member 4c | Q8R464 | Cadm4 | S,E |
| Neural cell adhesion molecule 1 | P13595 | Ncam1 | W,S,R,T,E |
| Neurofascin | Q810U3 | Nfasc | B,R,E |
| Oligodendrocyte myelin glycoprotein | Q63912 | Omg | ND |
| Opalin, TMP10 | Q7M750 | Opalin | R,E |
| Plasmolipin | Q9DCU2 | Pllp | E |
| Ras-related protein Rab 3A | P63011 | Rab3a | E |
| Ras-related protein Rab 3C | P62823 | Rab3c | E |
| Sirtuin 2 | Q8VDQ8 | Sirt2 | W,S,V,R,T,E |
| Tetraspanin 2 | Q922J6 | Tspan2 | E |
| B: Newly identified myelin-associated proteins | |||
| 14-3-3 protein beta | Q9CQV8 | Ywhab | E |
| 14-3-3 protein epsilon | P62259 | Ywhae | S,R,E |
| 14-3-3 protein eta | P68510 | Ywhah | E |
| 14-3-3 protein gamma | P61982 | Ywhag | W,V,R,T,E |
| 14-3-3 protein sigma, stratifin | O70456 | Sfn | E |
| 14-3-3 protein theta | P68254 | Ywhaq | E |
| 14-3-3 protein zeta delta | P63101 | Ywhaz | S,R,E |
| Actin α cardiac muscle 1 | P68033 | Actc1 | E |
| Actin α1 | P68134 | Acta1 | E |
| Actin α | P62737 | Acta2 | R,E |
| Actin β | P60710 | Actb | W,S,V,R,T,E |
| Actin γ1 | P63260 | Actg1 | B,E |
| Actin γ2 | P63268 | Actg2 | E |
| Acyl-CoA thioesterase 7 | Q91V12 | Acot7 | R,E |
| ADAM 23 | Q9R1V7 | Adam23 | E |
| Adenylate cyclase associated 1 | P40124 | Cap1 | T |
| ADP ribosylation factor 1 | P84078 | Arf1 | S,T,E |
| ADP ribosylation factor 2 | Q8BSL7 | Arf2 | E |
| ADP ribosylation factor 3 | P61205 | Arf3 | E |
| ADP ribosylation factor 4 | P61750 | Arf4 | E |
| ADP ribosylation factor 5 | P84084 | Arf5 | E |
| ADP ribosylation factor 6 | P62331 | Arf6 | W,E |
| Aldehyde dehydrogenase 1A1 | P24549 | Aldh1a1 | E |
| Aldolase A, fructose-bisphosphate | P05064 | Aldoa | W,S,V,R,T,E |
| Aldolase C, fructose bisphosphate | P05063 | Aldoc | R,T,E |
| Amphiphysin 2, bridging integrator 1 | O08539 | Bin1 | E |
| Anillin | Q8K298 | Anln | R,E |
| Annexin A2 | P07356 | Anxa2 | E |
| Annexin A6 | P14824 | Anxa6 | R,T |
| Argininosuccinate synthase 1 | P16460 | Ass1 | B,E |
| α-Synuclein | O55042 | Snca | E |
| Band 4.1 like protein 3 | Q9WV92 | Epb4.1l3 | E |
| Brain acid soluble protein 1, NAP22 | Q91XV3 | Basp1 | S,E |
| Breast carcinoma amplified seq 1 | Q80YN3 | Bcas1 | S,E |
| β-Synuclein | Q91ZZ3 | Sncb | E |
| Ca++ ATPase 1 | Q3TSK3 | Atp2b1 | E |
| Ca++ ATPase 2 | Q9R0K7 | Atp2b2 | E |
| Ca++ ATPase 3 | Q0VF55 | Atp2b3 | E |
| Ca++ ATPase 4 | Q6Q476 | Atp2b4 | E |
| Calmodulin CaM | P62204 | Calm3 | S,V,E |
| Calnexin | P35564 | Canx | B,R |
| Calpain 5 | O08688 | Capn5 | T |
| CaM kinase IIα | P11798 | Camk2a | E |
| CaM kinase IIβ | P28652 | Camk2b | E |
| CaM kinase IIδ | Q6PHZ2 | Camk2d | E |
| CaM kinase IIγ | Q923T9 | Camk2g | E |
| Cannabinoid receptor interacting 1 | Q5M8N0 | Cnrip1 | W,E |
| Carbonic anhydrase 2 | P00920 | Car2 | W,S,T,E |
| CD47, integrin signal transducer | Q61735 | Cd47 | E |
| CD82 | P40237 | Cd82 | E |
| CDGSH iron sulfur domain 1 | Q91WS0 | Cisd1 | E |
| Cell cycle exit and neuronal diff. | Q9JKC6 | Cend1 | E |
| Cell division control protein 42 | P60766 | Cdc42 | W,E |
| Centractin α | P61164 | Actr1a | W |
| Choline transporter CD92 | Q6X893 | Slc44a1 | E |
| Clathrin heavy chain | Q68FD5 | Cltc | B,R,E |
| Cofilin 1 | P18760 | Cfl1 | S,V,T,E |
| Cofilin 2 | P45591 | Cfl2 | E |
| Contactin associated protein 1 | O54991 | Cntnap1 | B,E |
| Coronin 1C | Q9WUM4 | Coro1c | E |
| Creatine kinase brain | Q04447 | Ckb | W,S,V,R,T,E |
| Crystallin α2 | P23927 | Cryab | W,S,T,E |
| Cyclophilin A | P17742 | Ppia | W,S,V,E |
| Cysteine and glycine rich protein 1 | P97315 | Csrp1 | E |
| Cytokeratin 1 | P04104 | Krt1 | E |
| Cytokeratin 1B | Q6IFZ6 | Krt77 | E |
| Cytokeratin 5 | Q922U2 | Krt5 | E |
| Cytokeratin 6A | P50446 | Krt6a | E |
| Cytokeratin 6G | Q9R0H5 | Krt71 | E |
| Cytokeratin 10 | P02535 | Krt10 | R,E |
| Cytokeratin 16 | Q9Z2K1 | Krt16 | E |
| Desmin | P31001 | Des | E |
| Destrin | Q9R0P5 | Dstn | E |
| Dihydropyrimidinase-like 1,CRMP1 | P97427 | Crmp1 | E |
| Dihydropyrimidinase-like 2,CRMP2 | O08553 | Dpysl2 | W,B,S,V,R,T,E |
| Dihydropyrimidinase-like 3,CRMP4 | Q62188 | Dpysl3 | E |
| Dihydropyrimidinase-like 4,CRMP3 | O35098 | Dpysl4 | E |
| Dipeptidylpeptidase 6 | Q9Z218 | Dpp6 | T |
| Down syndrome cell adhesion like 1 | Q8R4B4 | Dscaml1 | E |
| Dynactin 2 | Q99KJ8 | Dctn2 | V |
| Dynamin 1 | P39053 | Dnm1 | W,B,R,T,E |
| Dynamin 2 | P39054 | Dnm2 | E |
| Dynamin 3 | Q8BZ98 | Dnm3 | R |
| Dynein heavy chain | Q9JHU4 | Dync1h1 | R |
| Ectonucleotide pyrophosphatase 6 | Q8BGN3 | Enpp6 | E |
| EH domain containing protein 1 | Q9WVK4 | Ehd1 | B,S,T,E |
| EH domain containing protein 3 | Q9QXY6 | Ehd3 | B,E |
| EH domain containing protein 4 | Q9EQP2 | Ehd4 | E |
| Elongation factor 1α1 | P10126 | Eef1a1 | W,B,S,R,E |
| Elongation factor 1α2 | P62631 | Eef1a2 | W,B,E |
| Elongation factor 1β | O70251 | Eef1b2 | T |
| Elongation factor 2 | P58252 | Eef2 | T |
| Endonuclease domain containing 1 | Q8C522 | Endod1 | E |
| Enolase 1, non-neuronal | P17182 | Eno1 | W,B,S,V,T,E |
| Enolase 2, neuronal | P17183 | Eno2 | W,S,V,T,E |
| Enolase 3, muscle | P21550 | Eno3 | E |
| Fascin | Q61553 | Fscn1 | W,E |
| Fatty acid synthase | P19096 | Fasn | R |
| FK506 binding protein 1a | P26883 | Fkbp1a | S,E |
| Flotillin 1 | O08917 | Flot1 | ND, T (blot) |
| G protein α transducing 1 | P20612 | Gnat1 | E |
| G protein α transducing 2 | P50149 | Gnat2 | E |
| G protein α transducing 3 | Q3V3I2 | Gnat3 | E |
| G protein α11 | P21278 | Gna11 | E |
| G protein α14 | P30677 | Gna14 | E |
| G protein αI1 | B2RSH2 | Gnai1 | E |
| G protein αI2 | P08752 | Gnai2 | E |
| G protein αI3 | Q9DC51 | Gnai3 | E |
| G protein αO1 | P18872 | Gnao1 | S,T,E |
| G protein αO2 | P18873 | Gna0 | B,T,E |
| G protein αq | P21279 | Gnaq | T,E |
| G protein αS | P63094 | Gnas | S,E |
| G protein αS olfactory | Q8CGK7 | Gnal | E |
| G protein β1 | P62874 | Gnb1 | W,S,V,T,E |
| G protein β2 | P62880 | Gnb2 | W,V,R,E |
| G protein β3 | Q61011 | Gnb3 | E |
| G protein β4 | P29387 | Gnb4 | W,E |
| G protein β5 | P62881 | Gnb5 | W |
| G protein γ12 | Q9DAS9 | Gng12 | E |
| GAPDH | P16858 | Gapdh | W,S,V,T,E |
| GAPDH sperm | Q64467 | Gapdhs | E |
| Gelsolin | P13020 | Gsn | V,R,T |
| Glial fibrillary acidic protein | P03995 | Gfap | W,B |
| Glucose-6-phosphate isomerase | P06745 | Gpi1 | B,R,E |
| Glutamate oxaloacetate transaminase | P05201 | Got1 | E |
| Glutamate transporter GLAST | P56564 | Slc1a3 | E |
| Glutamate transporter GLT1 | P43006 | Slc1a2 | R,E |
| Glutamine synthetase | P15105 | Glul | W,S,V,T,E |
| Glutathione S transferase micros. 3 | Q9CPU4 | Mgst3 | E |
| Glutathione S transferase Mu1 | P10649 | Gstm1 | W,E |
| Glutathione S transferase Mu2 | P15626 | Gstm2 | E |
| Glutathione S transferase Mu6 | O35660 | Gstm6 | E |
| Glutathione S transferase P1 | P19157 | Gstp1 | S,V,E |
| Glutathione S transferase P2 | P46425 | Gstp2 | T |
| Growth associated protein 43 | P06837 | Gap43 | T |
| GTPase Ran | P62827 | Ran | E |
| H+/K+ ATPase α1 | Q64436 | Atp4a | E |
| H+/K+ ATPase α2 | Q9Z1W8 | Atp12a | E |
| Heat shock 70 kDa protein 1A | Q61696 | Hspa1a | E |
| Heat shock 70 kDa protein 1B | P17879 | Hspa1b | R,E |
| Heat shock 70 kDa protein 1L | P16627 | Hspa1l | E |
| Heat shock 70 kDa protein 2 | P17156 | Hspa2 | W,B,E |
| Heat shock 70 kDa protein 4 | Q61316 | Hspa4 | T |
| Heat shock 70 kDa protein 5 | P20029 | Hspa5 | W,T,E |
| Heat shock 70 kDa protein 8 | P63017 | Hspa8 | W,B,S,V,R,T,E |
| Heat shock 70 kDa protein 12A | Q8K0U4 | Hspa12a | E |
| Heat shock protein 90 kDa αA1 | P07901 | Hsp90aa1 | B,E |
| Heat shock protein 90 kDa αB1 | P11499 | Hsp90ab1 | T,E |
| Hexokinase 1 | P17710 | Hk1 | T,E |
| Hexokinase 2 | O08528 | Hk2 | E |
| Ig superfamily member 8, EWI-2 | Q8R366 | Igsf8 | B,S,R,E |
| Internexin α, Neurofilament 66 kDa | P46660 | Ina | W,B,V,R,T,E |
| Junctional adhesion molecule C | Q9D8B7 | Jam3 | S,E |
| K+ channel A1 | P16388 | Kcna1 | E |
| K+ channel A2 | P63141 | Kcna2 | E |
| K+ channel A3 | P16390 | Kcna3 | E |
| K+ channel B2 | P62482 | Kcnab2 | E |
| Lactate dehydrogenase A | P06151 | Ldha | T,E |
| Lactate dehydrogenase B | P16125 | Ldhb | W,T,E |
| Lactate dehydrogenase C | P00342 | Ldhc | E |
| Leucine rich repeat containing 57 | Q9D1G5 | Lrrc57 | E |
| Leucine rich repeat LGI 3 | Q8K406 | Lgi3 | E |
| Limbic system associated membrane | Q8BLK3 | Lsamp | S,E |
| Lymphocyte antigen 6H | Q9WUC3 | Ly6h | E |
| Macrophage migration inhibitory factor | P34884 | Mif | W,S,E |
| Malate dehydrogenase | P14152 | Mdh1 | W,S,V,T,E |
| MARCKS related protein | P28667 | Marcksl1 | S |
| Microtubule associated protein 1B | P14873 | Mtap1b | E |
| Microtubule associated protein 6 | Q7TSJ2 | Mtap6 | E |
| Microtubule associated protein tau | P10637 | Mapt | E |
| Mitogen activated protein kinase 1 | P63085 | Mapk1 | E |
| Moesin | P26041 | Msn | W,E |
| Munc 18, syntaxin binding protein 1 | O08599 | Stxbp1 | B,R,T,E |
| Myosin Id | Q5SYD0 | Myo1d | B,R,E |
| Na+/K+ ATPase α1 | Q8VDN2 | Atp1a1 | B,S,R,E |
| Na+/K+ ATPase α2 | Q6PIE5 | Atp1a2 | B,R,E |
| Na+/K+ ATPase α3 | Q6PIC6 | Atp1a3 | B,R,E |
| Na+/K+ ATPase α4 | Q9WV27 | Atp1a4 | E |
| Na+/K+ ATPase β1 | P14094 | Atp1b1 | B,S,R,E |
| Na+/K+ ATPase β3 | P97370 | Atp1b3 | E |
| Na+/K+/Cl− cotransporter | P55012 | Slc12a2 | E |
| N-ethylmaleimide sensitive fusion | P46460 | Nsf | W,B,R,T,E |
| Neurocalcin δ | Q91X97 | Ncald | S |
| Neurofilament H | P19246 | Nefh | W,B,E |
| Neurofilament L | P08551 | Nefl | W,B,V,R,E |
| Neurofilament M | P08553 | Nefm | B,R,E |
| Neuroligin 1 | Q99K10 | Nlgn1 | T |
| Neurotrimin | Q99PJ0 | Hnt | E |
| N-myc downstream regulated | Q62433 | Ndrg1 | W,S,V,T,E |
| Nucleoside diphosphate kinase A | P15532 | Nme1 | W,S,T,E |
| Nucleoside diphosphate kinase B | Q01768 | Nme2 | W,S,T,E |
| Parkinson disease protein 7 | Q99LX0 | Park7 | E |
| Peroxiredoxin 1 | P35700 | Prdx1 | W,V,R,T,E |
| Peroxiredoxin 2 | Q61171 | Prdx2 | W,V,E |
| Peroxiredoxin 5 | P99029 | Prdx5 | S,E |
| Phosphatidylethanolamine binding 1 | P70296 | Pebp1 | W,V,E |
| Phosphatidylinositol transfer α | P53810 | Pitpna | W |
| Phosphofructokinase 1 | P47857 | Pfkm | E |
| Phosphoglycerate dehydrogenase | Q61753 | Phgdh | W |
| Phosphoglycerate kinase 1 | P09411 | Pgk1 | S,V,T,E |
| Phosphoglycerate kinase 2 | P09041 | Pgk2 | E |
| Phosphoglycerate mutase 1 | Q9DBJ1 | Pgam1 | W,S,T,E |
| Phospholipase Cβ1 | Q9Z1B3 | Plcb1 | W,T,E |
| Phosphoserine aminotransferase | Q99K85 | Psat1 | R |
| Prion protein | P04925 | Prnp | E |
| Prion protein dublet | Q9QYT9 | Prnd | E |
| Programmed cell death 6 interacting | Q9WU78 | Pdcd6ip | W |
| Prohibitin | P67778 | Phb | W,B,E |
| Prohibitin 2 | O35129 | Phb2 | E |
| Protein arginine deiminase 2 | Q08642 | Padi2 | E |
| Protein disulfide isomerase A3 | P27773 | Pdia3 | W,T |
| Protein kinase Cγ | P63318 | Prkcc | E |
| Pyruvate kinase isozyme M2 | P52480 | Pkm2 | W,S,V,T,E |
| Quinoid dihydropteridine reductase | Q8BVI4 | Qdpr | E |
| Rab 1A | P62821 | Rab1 | E |
| Rab 1B | Q9D1G1 | Rab1b | E |
| Rab 2A | P53994 | Rab2a | R,E |
| Rab 2B | P59279 | Rab2b | E |
| Rab 3B | Q9CZT8 | Rab3b | E |
| Rab 3D | P35276 | Rab3d | E |
| Rab 4A | P56371 | Rab4a | E |
| Rab 4B | Q91ZR1 | Rab4b | E |
| Rab 5C | P35278 | Rab5c | E |
| Rab 7A | P51150 | Rab7 | R |
| Rab 8A | P55258 | Rab8a | E |
| Rab 8B | P61028 | Rab8b | E |
| Rab 10 | P61027 | Rab10 | S,E |
| Rab 12 | P35283 | Rab12 | E |
| Rab 13 | Q9DD03 | Rab13 | E |
| Rab 14 | Q91V41 | Rab14 | E |
| Rab 15 | Q8K386 | Rab15 | E |
| Rab 18 | P35293 | Rab18 | E |
| Rab 26 | Q504M8 | Rab26 | E |
| Rab 30 | Q923S9 | Rab30 | E |
| Rab 35 | Q6PHN9 | Rab35 | E |
| Rab 37 | Q9JKM7 | Rab37 | E |
| Rab 39B | Q8BHC1 | Rab39b | E |
| Rab 43 | Q8CG50 | Rab43 | E |
| Rab GDP dissociation inhibitor α | P50396 | Gdi1 | W,S,R,T,E |
| Rab GDP dissociation inhibitor β | Q61598 | Gdi2 | W,T,E |
| Rac1 | P63001 | Rac1 | S,R,E |
| Rac2 | Q05144 | Rac2 | E |
| Rac3 | P60764 | Rac3 | E |
| Radixin | P26043 | Rdx | W,E |
| Ras-related protein Ral A | P63321 | Rala | B,E |
| Ras-related protein Ral B | Q9JIW9 | Ralb | E |
| Ras-related protein Rap 1A | P62835 | Rap1a | W,S,R,T,E |
| Ras-related protein Rap 1B | Q99JI6 | Rap1b | E |
| Ras-related protein Rap 2a | Q80ZJ1 | Rap2a | R |
| Reticulon 3 | Q9ES97 | Rtn3 | R |
| Rho GDP dissociation inhibitor 1 | Q99PT1 | Arhgdia | V,T |
| RhoA | Q9QUI0 | Rhoa | E |
| RhoB | P62746 | Rhob | T,E |
| RhoC | Q62159 | Rhoc | E |
| RhoG | P84096 | Rhog | E |
| S-100β | P50114 | S100b | R |
| Septin 2 | P42208 | Sept2 | W,B,S,T,E |
| Septin 4 | P28661 | Sept4 | W,E |
| Septin 7 | O55131 | Sept7 | W,B,S,R,T,E |
| Septin 8 | Q8CHH9 | Sept8 | W,B,S,V,R,T,E |
| Septin 11 | Q8C1B7 | Sept11 | E |
| Sideroflexin 3 | Q91V61 | Sfxn3 | E |
| Soluble NSF attachment protein α | Q9DB05 | Napa | W |
| Soluble NSF attachment protein β | P28663 | Napb | W,E |
| Soluble NSF attachment protein γ | Q9CWZ7 | Napg | W |
| Spectrin α2 | P16546 | Spna2 | B,T,E |
| Spectrin β2 | Q62261 | Spnb2 | R,E |
| Stress induced phosphoprotein 1 | Q60864 | Stip1 | W,T |
| Superoxide dismutase | P08228 | Sod1 | W,S |
| Synapsin 1 | O88935 | Syn1 | W,E |
| Synapsin 2 | Q64332 | Syn2 | W,E |
| Synaptic vesicle membrane protein | Q62465 | Vat1 | R,T |
| Synaptobrevin 2 | P63044 | Vamp2 | E |
| Synaptobrevin 3 | P63024 | Vamp3 | E |
| Synaptophysin | Q62277 | Syp | E |
| Synaptosomal associated protein 23 | O09044 | Snap23 | E |
| Synaptosomal associated protein 25 | P60879 | Snap25 | W,S,V,R,E |
| Synaptotagmin 1 | P46096 | Syt1 | E |
| Synaptotagmin 5 | Q9R0N5 | Syt5 | E |
| Syndapin 1 | Q61644 | Pacsin1 | W,E |
| Syntaxin 1A | O35526 | Stx1a | E |
| Syntaxin 1B | P61264 | Stx1b | S,R,E |
| T-complex 1α | P11983 | Tcp1 | W |
| T-complex 1β | P80314 | Cct2 | W |
| T-complex 1δ | P80315 | Cct4 | R |
| T-complex 1ε | P80316 | Cct5 | W |
| T-complex 1γ | P80318 | Cct3 | W |
| Thy 1 membrane glycoprotein | P01831 | Thy1 | W,S,R,E |
| Transgelin 3 | Q9R1Q8 | Tagln3 | W,E |
| Transitional ER ATPase | Q01853 | Vcp | W,T,E |
| Transketolase | P40142 | Tkt | W,B,S,T,E |
| Triosephosphate isomerase | P17751 | Tpi1 | S,E |
| Tubulin α1A | P68369 | Tuba1a | W,B,R,E |
| Tubulin α1B | P05213 | Tuba1b | W,S,V,T,E |
| Tubulin α1C | P68373 | Tuba1c | E |
| Tubulin α3A | P05214 | Tuba3a | E |
| Tubulin α4A | P68368 | Tuba4a | E |
| Tubulin α8 | Q9JJZ2 | Tuba8 | E |
| Tubulin β2A | Q7TMM9 | Tubb2a | T,E |
| Tubulin β2B | Q9CWF2 | Tubb2b | E |
| Tubulin β2C | P68372 | Tubb2c | W,B,S,R,E |
| Tubulin β3 | Q9ERD7 | Tubb3 | E |
| Tubulin β4 | Q9D6F9 | Tubb4 | W,B,S,V,R,E |
| Tubulin β5 | P99024 | Tubb5 | E |
| Tubulin β6 | Q922F4 | Tubb6 | R,E |
| Tubulin polymerization promoting | Q7TQD2 | Tppp | W,E |
| Tubulin polymerization promoting 3 | Q9CRB6 | Tppp3 | S,E |
| Ubiquitin | P62991 | Ub | W,S,E |
| Ubiquitin activating enzyme E1 | Q02053 | Uba1 | T |
| Ubiquitin C-terminal hydrolase L1 | Q9R0P9 | Uchl1 | W,T,E |
| Vacuolar ATP synthase A | P50516 | Atp6v1a | W,E |
| Vacuolar ATP synthase B, brain | P62814 | Atp6v1b2 | W,E |
| Vacuolar ATP synthase C | Q9Z1G3 | Atp6v1c1 | T,E |
| Vacuolar ATP synthase E1 | P50518 | Atp6v1e1 | T,E |
| Vimentin | P20152 | Vim | E |
| Visinin like protein 1 | P62761 | Vsnl1 | S,R,E |
| Visinin like protein 3 | P62748 | Hpcal1 | S |
| WD repeat protein 1 | O88342 | Wdr1 | W |
Proteins identified in purified CNS myelin by MS
ID Swissprot or Trembl accession, Gene official NCBI Entrez gene name, Reference and method of detection, T 2D-IEF/SDS-PAGE or immunoblotting [95], V 2D-IEF/SDS-PAGE [97], W 2D-IEF/SDS-PAGE [25], B 2D-16-BAC/SDS-PAGE [25], R shotgun [23], S shotgun [97], E LC-MSE (Tenzer et al., unpublished), ND not detected by MS
Fig. 3.
Assembling a compendium of myelin proteins. a The number of proteins identified by MS in different approaches to the CNS myelin proteome is plotted. The total number of myelin-associated proteins is unknown. Transmembrane proteins (black) have been categorized based on prior experimental studies or have been predicted using TMHMM and Phobius software. Proteins associated with mitochondria, which copurify with myelin, were omitted. T 2D-IEF/SDS-PAGE [95], V 2D-IEF/SDS-PAGE [97], W 2D-IEF/SDS-PAGE [25], B 2D-16-BAC/SDS-PAGE [25], R shotgun [23], S shotgun [97], E LC-MSE (Tenzer et al., unpublished). b Venn diagram comparing the number of myelin-associated proteins identified by MS after gel separation [25, 95, 97], previous gel-free shotgun approaches by LC/LC-MS/MS [23, 97], with those identified by LC-MSE (Tenzer et al., unpublished). Note the high overlap of proteins identified independent of the technique used. c Venn diagram showing our own experience with the identification of myelin-associated proteins by MS after combined 2D-IEF/SDS-PAGE and 2D-16-BAC/SDS-PAGE separation [25] or by LC-MSE with known myelin proteins according to the literature
Fig. 4.
Relative abundance of myelin proteins. a The abundance of known myelin proteins was determined by LC-MSE. Note that known myelin proteins constitute less than 50% of the total myelin protein. Mitochondrial proteins were not considered. b Comparison of myelin protein abundance as quantified by LC-MSE with previous estimates based on band intensity after 1D-PAGE and various protein staining techniques [19, 85, 87, 88]. Note that the abundance of PLP and MBP was previously overestimated because low abundant proteins did not constitute significant bands due to limitations in the resolving power of the 1D gels and in the dynamic range of protein staining. c Simulated 2D map of myelin-associated proteins identified by LC-MSE. Proteins are indicated as dots at their molecular weight and isoelectric point as predicted from the amino acid sequence. The size of each dot reflects the relative abundance as determined by LC-MSE. Myelin-associated proteins without transmembrane domains are shown in blue and transmembrane proteins in green, the latter being usually under-represented or absent from conventional 2D gels. Mitochondrial proteins are shown in gray. The red frame indicates the portion of proteins that can be reproducibly displayed by 2D-IEF/SDS-PAGE (see Fig. 2a)
We conclude that modern LC-MS-based approaches—though technically more demanding than gel-based studies—appear to be appropriate for tackling the myelin proteome as they cover several orders of magnitude of protein abundance and detect highly basic, hydrophobic, and membrane-spanning proteins. This tackles the bias towards certain protein classes, which is the major shortcoming particularly of 2D-IEF/SDS-PAGE (Fig. 4c). Moreover, LC-MS-based approaches enable the gel- and label-free quantification of proteins from complex mixtures, which allowed for the systematic reassignment of protein abundance in CNS myelin (see above). Finally, they require only low amounts of sample, which is of special relevance for the proteome analysis of myelin purified from hypomyelinated model animals or human brain autopsy material.
Technical Limitations
How pure is the myelin-enriched fraction? Myelin-associated proteins are defined as proteins in the myelin-enriched fraction since all studies have operationally defined the term “myelin protein” without systematic experimental verification. Although the identification of new myelin proteins by more than one study and the detection of established myelin markers increase confidence, some of these proteins may only have copurified with myelin. The high dynamic range of LC-MSE leads to the new identification of proteins as myelin-associated, but also to the false-positive identification of contaminants. These mainly stem from copurifying mitochondria and synaptic vesicles. In reverse, proteomic compendia of mammalian brain mitochondria [116] or synaptic vesicles [90] include classical myelin proteins such as PLP, MBP, MOBP, and MAG. Notwithstanding that some of these proteins may have a dual localization, cross-contamination occurs likely due to similar floatation properties in sucrose or Percoll gradients and can only be excluded once improved separation protocols become available. Proteins of the axonal plasma membrane, such as potassium channels or Na+/K+-ATPases, have also been detected in the myelin fraction, which can be explained by the tight linkage of the membranes via adhesion proteins, sometimes referred to as the myelin–axolemma complex [24]. Indeed, some adhesion complexes are present in the myelin-enriched fraction, such as the glial neurofascin (NF155) and contactin and their axonal partner contactin associated protein 1 (Caspr) [117–120] and the glial nectin-like protein Necl4 and its axonal counterpart Necl1 [121–124]. Importantly, myelin proteome analysis also revealed novel candidate proteins to mediate intracellular or intercellular adhesion, such as the immunoglobulin domain superfamily protein Igsf8, also termed EWI-2 [23]. Igsf8 is associated with the myelin tetraspanins CD9 and CD81 and regulates integrin function, at least in vitro [125, 126], but its function in vivo remains to be shown. The experimental validation or falsification of newly identified myelin-associated proteins will be a matter of the systematic application of histological techniques, provided that reliable antibodies are available.
How many proteins can be considered true myelin proteins? Though proteomic compendia aim at completeness, the number can only be guessed at this time. As the dynamic range of current MS-based protein identification schemes is in the range of three to five orders of magnitude, detection of infrequent proteins remains a challenge. Additionally, some technical impediments remain. The myelin proteins CD9 [127, 128], oligodendrocyte myelin glycoprotein [22, 129], and MAL [51] have not yet been detected by proteomic approaches, and the appearance of MAL in one catalog [95] is due to the additional use of immunoblotting. Its nondetectability illustrates the limitations of proteome analysis. MAL is a very hydrophobic protein with four transmembrane domains and very small cytoplasmic and extracellular domains and is, therefore, hardly accessible by MS-based identification. Apart from the membrane-spanning peptides not visible in proteomic approaches, complete tryptic digest of MAL results in only four theoretically detectable peptides: one of 120 amino acids (which is too long for identification by MS), two of two amino acids each (too short to provide useful sequence information), and one of 29 amino acids, which is, in principle, appropriate for identification. However, to obtain a reasonable level of confidence for protein identification, the detection of two peptides per protein is usually set as a prerequisite in the algorithms. This suggests that all proteome approaches requiring protease cleavage have an inherent bias against very small polypeptides or proteins with an unusual cleavage site pattern. In future experiments, the lack of suitable trypsin cleavage sites may be circumvented by the use of endopeptidases with different specificities (e.g., GluC or AspN), although they create proteolytic peptides lacking a basic C-terminal amino acid and are difficult to sequence [130]. This suggests that the detection of more myelin-associated proteins is not just a matter of higher resolving power but also of other technical refinements.
Newly Identified Myelin-Associated Proteins
The compendium of proteins identified in the myelin-enriched brain fraction represents a valuable reference for myelin research. The proteins are candidates for performing important functions in myelin biogenesis and integrity, molecular interactions between myelinating glia and neighboring cells, and white matter homeostasis. By gene ontology terms (http://david.abcc.ncifcrf.gov), many myelin-associated proteins are implicated in catalytic activities (48%), the cytoskeleton (20%), protein transport (21%), vesicular trafficking (6.8%), cell adhesion (6.3%), phospholipid binding (4.2%), or glycolysis/gluconeogenesis (5.1%). Among the recently identified myelin proteins, some were first and others subsequently detected using proteomic approaches. They include proteins of quite various anticipated functions, such as the NAD+-dependent deacetylase sirtuin 2 (SIRT2, see below), cytoskeletal proteins of the septin family [23, 25, 131], and ermin [132], regulators of intracellular vesicle transport in the secretory pathway, such as cdc42 and Rac1 [133], Rab3A, and other Rab-GTPases [134, 135], the paranodal transmembrane glycoprotein Opalin/TMEM10 with a suggested signaling or adhesive function [136–138], the nucleoside diphosphate kinases NM23A and NM23B [95], and a protein particularly abundant in the CNS myelin of teleost fish, the 36K protein, also termed short-chain dehydrogenase/reductase (SDR family) member 12 (DHRS12) [139]. Some of these are quite abundant myelin proteins as judged both by the spots constituted on 2D gels and LC-based quantification, and the challenge to establish their functions in vivo promises a deepened understanding of myelin. Besides, novel myelin proteins are candidates to cause (when mutated), enhance, or ameliorate white matter disease, such as leukodystrophies.
Differential Myelin Proteome Analysis in Myelin-Related Disease
The proteomic comparison of myelin from human patients or animal models with that of respective controls is a powerful approach towards the identification of secondary molecular changes that may contribute to the pathogenesis of myelin-related disease. Such a differential approach has first been applied to myelin purified from PLPnull mice [25], which provide a genuine model for spastic paraplegia (SPG-2) in humans, a comparatively mild variant of the leukodystrophy Pelizaeus–Merzbacher Disease with progressive axonal degeneration in the presence of normal amounts of CNS myelin [29, 140]. In that study, 2D-DIGE [141] was used to screen for candidate proteins that could be involved in the oligodendroglial failure to support the long-term integrity of myelinated axons. Three distinct proteins of the cytoskeletal septin family were found to be reduced, and the deacetylase SIRT2 was virtually absent from PLPnull myelin. SIRT2 is an abundant myelin protein in the CNS and the PNS [23, 25, 142] and regulates microtubule dynamics during oligodendrocyte development [143]. Whether acetylated α-tubulin is a relevant substrate of SIRT2 in vivo remains to be shown. Similar to PLPnull mice, CNPnull mice are also normally myelinated but develop length-dependent axonal loss [92, 144]. It is intriguing that CNP also modulates microtubule dynamics [145, 146]. Taken together, spatiotemporal control of microtubule stability in oligodendrocytes (by SIRT2, CNP, and likely other factors) seems critical for normal axon–glia interaction.
Acetylation is a reversible post-translational modification of numerous mammalian proteins [147, 148], and all acetylated myelin proteins (α-tubulin, MBP, MOG, and several nonidentified proteins of lower abundance) are candidate substrates for SIRT2 [25]. In oligodendrocytes and myelin, SIRT2 activation upon increased axonal NAD+ levels may remove acetyl residues from myelin-associated proteins with consequences for their net charge and function. Interestingly, SIRT2 has been recently shown to interact with 14-3-3 beta and gamma [149], which are myelin-associated as revealed by proteome analysis (Table 1). Their interaction is strengthened by the serine/threonine kinase AKT [149], which is a central signaling molecule for CNS myelination [150]. 14-3-3 proteins have been implicated in membrane protein transport, exocytosis [151], and stress response [152], but their function in myelin has not yet been investigated. 14-3-3 proteins are homologs of the C. elegans partitioning-defective polarity protein Par5 and bind to the tight junction-associated Par3 [153, 154], which is required for establishing polarity prior to myelination, at least by Schwann cells in the PNS [155]. To determine whether SIRT2, 14-3-3 proteins, Par-proteins, protein kinases, and tight junctions indeed interact in myelinating glia will be an important topic of future investigation. We speculate that the competence of oligodendrocytes to dynamically react to NAD+ level changes in white matter tracts is required for their role in maintaining long-term axonal integrity.
With the objective to identify novel therapeutic targets for the treatment of multiple sclerosis, a systematic proteomic profiling of tissue samples from three brain lesions affected to various degrees (acute plaque, chronic active plaque, and chronic plaque) has recently been performed [156]. Material from the respective lesion type was collected by laser-capture microdissection and extracted proteins were separated by 1D gel electrophoresis followed by mass spectrometric protein identification. Unexpectedly, five coagulation proteins, including tissue factor and protein C inhibitor, were only present in chronic active plaque characterized by concomitant inflammation and degeneration, a finding that provided new insights in the relationship between the coagulation cascade and inflammation. Most importantly, administration of inhibitors to tissue factor (i.e., hirudin) and protein C inhibitor (i.e., activated protein C [aPC]) indeed ameliorated the disease phenotype in experimental autoimmune encephalomyelitis, a model of multiple sclerosis. The anti-inflammatory treatment with engineered aPC variants may develop into an alternative route to a therapy of multiple sclerosis. Together, differential proteome analysis has identified secondary molecular changes that contribute to understanding the pathogenesis of myelin-related disease and support the design of rational treatment strategies.
Acknowledgements
We thank S. Wichert, W. Möbius, J. Patzig, I. Ionescu, and K.-A. Nave for the discussions. ST is supported by the Deutsche Forschungsgemeinschaft (SFB 490 Z3) and the Forschungszentrum Immunologie (FZI) at the University of Mainz, and HW is supported by the BMBF (DLR-Leukonet).
References
- 1.Hartline DK, Colman DR (2007) Rapid conduction and the evolution of giant axons and myelinated fibers. Curr Biol 17:R29–R35 [DOI] [PubMed]
- 2.Poliak S, Peles E (2003) The local differentiation of myelinated axons at nodes of Ranvier. Nat Rev Neurosci 4:968–980 [DOI] [PubMed]
- 3.Hildebrand C, Remahl S, Persson H, Bjartmar C (1993) Myelinated nerve fibres in the CNS. Prog Neurobiol 40:319–384 [DOI] [PubMed]
- 4.Kirby BB, Takada N, Latimer AJ, Shin J, Carney TJ, Kelsh RN, Appel B (2006) In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat Neurosci 9:1506–1511 [DOI] [PubMed]
- 5.Brosamle C, Halpern ME (2002) Characterization of myelination in the developing zebrafish. Glia 39:47–57 [DOI] [PubMed]
- 6.Pogoda HM, Sternheim N, Lyons DA, Diamond B, Hawkins TA, Woods IG, Bhatt DH, Franzini-Armstrong C, Dominguez C, Arana N, Jacobs J, Nix R, Fetcho JR, Talbot WS (2006) A genetic screen identifies genes essential for development of myelinated axons in zebrafish. Dev Biol 298:118–131 [DOI] [PubMed]
- 7.Schweitzer J, Becker T, Schachner M, Nave KA, Werner H (2006) Evolution of myelin proteolipid proteins: gene duplication in teleosts and expression pattern divergence. Mol Cell Neurosci 31:161–177 [DOI] [PubMed]
- 8.Avila RL, Tevlin BR, Lees JP, Inouye H, Kirschner DA (2007) Myelin structure and composition in zebrafish. Neurochem Res 32:197–209 [DOI] [PubMed]
- 9.Sherman DL, Brophy PJ (2005) Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci 6:683–690 [DOI] [PubMed]
- 10.Simons M, Trotter J (2007) Wrapping it up: the cell biology of myelination. Curr Opin Neurobiol 17:533–540 [DOI] [PubMed]
- 11.Nave KA, Trapp BD (2008) Axon-glial signaling and the glial support of axon function. Annu Rev Neurosci 31:535–561 [DOI] [PubMed]
- 12.Lassmann H, Lucchinetti CF (2008) Cortical demyelination in CNS inflammatory demyelinating diseases. Neurology 70:332–333 [DOI] [PubMed]
- 13.Boespflug-Tanguy O, Labauge P, Fogli A, Vaurs-Barriere C (2008) Genes involved in leukodystrophies: a glance at glial functions. Curr Neurol Neurosci Rep 8:217–229 [DOI] [PubMed]
- 14.Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA (2006) Functional genomic analysis of oligodendrocyte differentiation. J Neurosci 26:10967–10983 [DOI] [PMC free article] [PubMed]
- 15.Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD (2006) Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci 26:9881–9891 [DOI] [PMC free article] [PubMed]
- 16.Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, Barres BA (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci 28:264–278 [DOI] [PMC free article] [PubMed]
- 17.Dumont D, Noben JP, Moreels M, Vanderlocht J, Hellings N, Vandenabeele F, Lambrichts I, Stinissen P, Robben J (2007) Characterization of mature rat oligodendrocytes: a proteomic approach. J Neurochem 102:562–576 [DOI] [PubMed]
- 18.Krämer-Albers E-M, Bretz N, Tenzer S, Winterstein C, Möbius W, Berger H, Nave K-A, Schild H, Trotter J (2007) Oligodendrocytes secrete exosomes containing major myelin and stress-protective proteins: trophic support for axons? Proteomics Clin Appl 1:1446–1461 [DOI] [PubMed]
- 19.Norton WT, Poduslo SE (1973) Myelination in rat brain: method of myelin isolation. J Neurochem 21:749–757 [DOI] [PubMed]
- 20.Larocca JN, Norton WT (2007) Isolation of myelin. Curr Protoc Cell Biol Chapter 3:Unit3.25 [DOI] [PubMed]
- 21.Colman DR, Kreibich G, Frey AB, Sabatini DD (1982) Synthesis and incorporation of myelin polypeptides into CNS myelin. J Cell Biol 95:598–608 [DOI] [PMC free article] [PubMed]
- 22.Huang JK, Phillips GR, Roth AD, Pedraza L, Shan W, Belkaid W, Mi S, Fex-Svenningsen A, Florens L, Yates JR 3rd, Colman DR (2005) Glial membranes at the node of Ranvier prevent neurite outgrowth. Science 310:1813–1817 [DOI] [PubMed]
- 23.Roth AD, Ivanova A, Colman DR (2006) New observations on the compact myelin proteome. Neuron Glia Biol 2:15–21 [DOI] [PubMed]
- 24.Menon K, Rasband MN, Taylor CM, Brophy P, Bansal R, Pfeiffer SE (2003) The myelin-axolemmal complex: biochemical dissection and the role of galactosphingolipids. J Neurochem 87:995–1009 [DOI] [PubMed]
- 25.Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Mobius W, Guarente L, Casaccia-Bonnefil P, Jahn O, Nave KA (2007) Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci 27:7717–7730 [DOI] [PMC free article] [PubMed]
- 26.Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, Wieland F, Ishibashi S, Nave KA (2005) High cholesterol level is essential for myelin membrane growth. Nat Neurosci 8:468–475 [DOI] [PubMed]
- 27.Coetzee T, Fujita N, Dupree J, Shi R, Blight A, Suzuki K, Popko B (1996) Myelination in the absence of galactocerebroside and sulfatide: normal structure with abnormal function and regional instability. Cell 86:209–219 [DOI] [PubMed]
- 28.Saravanan K, Schaeren-Wiemers N, Klein D, Sandhoff R, Schwarz A, Yaghootfam A, Gieselmann V, Franken S (2004) Specific downregulation and mistargeting of the lipid raft-associated protein MAL in a glycolipid storage disorder. Neurobiol Dis 16:396–406 [DOI] [PubMed]
- 29.Klugmann M, Schwab MH, Puhlhofer A, Schneider A, Zimmermann F, Griffiths IR, Nave KA (1997) Assembly of CNS myelin in the absence of proteolipid protein. Neuron 18:59–70 [DOI] [PubMed]
- 30.Norton WT, Poduslo SE (1973) Myelination in rat brain: changes in myelin composition during brain maturation. J Neurochem 21:759–773 [DOI] [PubMed]
- 31.Morell P, Jurevics H (1996) Origin of cholesterol in myelin. Neurochem Res 21:463–470 [DOI] [PubMed]
- 32.Huttner WB, Zimmerberg J (2001) Implications of lipid microdomains for membrane curvature, budding and fission. Curr Opin Cell Biol 13:478–484 [DOI] [PubMed]
- 33.Bosio A, Binczek E, Haupt WF, Stoffel W (1998) Composition and biophysical properties of myelin lipid define the neurological defects in galactocerebroside- and sulfatide-deficient mice. J Neurochem 70:308–315 [DOI] [PubMed]
- 34.Dupree JL, Coetzee T, Blight A, Suzuki K, Popko B (1998) Myelin galactolipids are essential for proper node of Ranvier formation in the CNS. J Neurosci 18:1642–1649 [DOI] [PMC free article] [PubMed]
- 35.Honke K, Hirahara Y, Dupree J, Suzuki K, Popko B, Fukushima K, Fukushima J, Nagasawa T, Yoshida N, Wada Y, Taniguchi N (2002) Paranodal junction formation and spermatogenesis require sulfoglycolipids. Proc Natl Acad Sci U S A 99:4227–4232 [DOI] [PMC free article] [PubMed]
- 36.Ishibashi T, Dupree JL, Ikenaka K, Hirahara Y, Honke K, Peles E, Popko B, Suzuki K, Nishino H, Baba H (2002) A myelin galactolipid, sulfatide, is essential for maintenance of ion channels on myelinated axon but not essential for initial cluster formation. J Neurosci 22:6507–6514 [DOI] [PMC free article] [PubMed]
- 37.Hirahara Y, Bansal R, Honke K, Ikenaka K, Wada Y (2004) Sulfatide is a negative regulator of oligodendrocyte differentiation: development in sulfatide-null mice. Glia 45:269–277 [DOI] [PubMed]
- 38.Zoller I, Meixner M, Hartmann D, Bussow H, Meyer R, Gieselmann V, Eckhardt M (2008) Absence of 2-hydroxylated sphingolipids is compatible with normal neural development but causes late-onset axon and myelin sheath degeneration. J Neurosci 28:9741–9754 [DOI] [PMC free article] [PubMed]
- 39.Eckhardt M, Hedayati KK, Pitsch J, Lullmann-Rauch R, Beck H, Fewou SN, Gieselmann V (2007) Sulfatide storage in neurons causes hyperexcitability and axonal degeneration in a mouse model of metachromatic leukodystrophy. J Neurosci 27:9009–9021 [DOI] [PMC free article] [PubMed]
- 40.Ramakrishnan H, Hedayati KK, Lullmann-Rauch R, Wessig C, Fewou SN, Maier H, Goebel HH, Gieselmann V, Eckhardt M (2007) Increasing sulfatide synthesis in myelin-forming cells of arylsulfatase A-deficient mice causes demyelination and neurological symptoms reminiscent of human metachromatic leukodystrophy. J Neurosci 27:9482–9490 [DOI] [PMC free article] [PubMed]
- 41.Eckhardt M (2008) The role and metabolism of sulfatide in the nervous system. Mol Neurobiol 37:93–103 [DOI] [PubMed]
- 42.Farooqui AA, Horrocks LA (2006) Phospholipase A2-generated lipid mediators in the brain: the good, the bad, and the ugly. Neuroscientist 12:245–260 [DOI] [PubMed]
- 43.Kassmann CM, Nave KA (2008) Oligodendroglial impact on axonal function and survival—a hypothesis. Curr Opin Neurol 21:235–241 [DOI] [PubMed]
- 44.Brosche T, Platt D (1998) The biological significance of plasmalogens in defense against oxidative damage. Exp Gerontol 33:363–369 [DOI] [PubMed]
- 45.Brites P, Mooyer PA, El Mrabet L, Waterham HR, Wanders RJ (2008) Plasmalogens participate in very-long-chain fatty acid-induced pathology. Brain 132:482–492 [DOI] [PubMed]
- 46.Khan M, Singh J, Singh I (2008) Plasmalogen deficiency in cerebral adrenoleukodystrophy and its modulation by lovastatin. J Neurochem 106:1766–1779 [DOI] [PMC free article] [PubMed]
- 47.Fourcade S, Lopez-Erauskin J, Galino J, Duval C, Naudi A, Jove M, Kemp S, Villarroya F, Ferrer I, Pamplona R, Portero-Otin M, Pujol A (2008) Early oxidative damage underlying neurodegeneration in X-adrenoleukodystrophy. Hum Mol Genet 17:1762–1773 [DOI] [PubMed]
- 48.Hein S, Schonfeld P, Kahlert S, Reiser G (2008) Toxic effects of X-linked adrenoleukodystrophy-associated, very long chain fatty acids on glial cells and neurons from rat hippocampus in culture. Hum Mol Genet 17:1750–1761 [DOI] [PubMed]
- 49.Rodemer C, Thai TP, Brugger B, Kaercher T, Werner H, Nave KA, Wieland F, Gorgas K, Just WW (2003) Inactivation of ether lipid biosynthesis causes male infertility, defects in eye development and optic nerve hypoplasia in mice. Hum Mol Genet 12:1881–1895 [DOI] [PubMed]
- 50.Frank M, Schaeren-Wiemers N, Schneider R, Schwab ME (1999) Developmental expression pattern of the myelin proteolipid MAL indicates different functions of MAL for immature Schwann cells and in a late step of CNS myelinogenesis. J Neurochem 73:587–597 [DOI] [PubMed]
- 51.Schaeren-Wiemers N, Bonnet A, Erb M, Erne B, Bartsch U, Kern F, Mantei N, Sherman D, Suter U (2004) The raft-associated protein MAL is required for maintenance of proper axon–glia interactions in the central nervous system. J Cell Biol 166:731–742 [DOI] [PMC free article] [PubMed]
- 52.Folch J, Lees M (1951) Proteolipides, a new type of tissue lipoproteins; their isolation from brain. J Biol Chem 191:807–817 [PubMed]
- 53.Lees MB (1998) A history of proteolipids: a personal memoir. Neurochem Res 23:261–271 [DOI] [PubMed]
- 54.Brophy PJ, Horvath LI, Marsh D (1984) Stoichiometry and specificity of lipid-protein interaction with myelin proteolipid protein studied by spin-label electron spin resonance. Biochemistry 23:860–865 [DOI] [PubMed]
- 55.Swamy MJ, Horvath LI, Brophy PJ, Marsh D (1999) Interactions between lipid-anchored and transmembrane proteins. Spin-label ESR studies on avidin-biotinyl phosphatidylethanolamine in membrane recombinants with myelin proteolipid proteins. Biochemistry 38:16333–16339 [DOI] [PubMed]
- 56.Simons M, Kramer EM, Thiele C, Stoffel W, Trotter J (2000) Assembly of myelin by association of proteolipid protein with cholesterol- and galactosylceramide-rich membrane domains. J Cell Biol 151:143–154 [DOI] [PMC free article] [PubMed]
- 57.Kramer-Albers EM, Gehrig-Burger K, Thiele C, Trotter J, Nave KA (2006) Perturbed interactions of mutant proteolipid protein/DM20 with cholesterol and lipid rafts in oligodendroglia: implications for dysmyelination in spastic paraplegia. J Neurosci 26:11743–11752 [DOI] [PMC free article] [PubMed]
- 58.Folch J, Lebaron FN (1956) The isolation from brain tissue of a trypsin-resistant protein fraction containing combined inositol, and its relation to neurokeratin. J Neurochem 1:101–108 [DOI] [PubMed]
- 59.Lees MB, Leston JA, Paxman SA (1971) The heterogeneity of the trypsin-resistant protein residue from brain white matter. J Neurochem 18:1791–1794 [DOI] [PubMed]
- 60.Gielen E, Baron W, Vandeven M, Steels P, Hoekstra D, Ameloot M (2006) Rafts in oligodendrocytes: evidence and structure–function relationship. Glia 54:499–512 [DOI] [PubMed]
- 61.Debruin LS, Harauz G (2007) White matter rafting—membrane microdomains in myelin. Neurochem Res 32:213–228 [DOI] [PubMed]
- 62.de Vries H, Hoekstra D (2000) On the biogenesis of the myelin sheath: cognate polarized trafficking pathways in oligodendrocytes. Glycoconj J 17:181–190 [DOI] [PubMed]
- 63.Kramer EM, Schardt A, Nave KA (2001) Membrane traffic in myelinating oligodendrocytes. Microsc Res Tech 52:656–671 [DOI] [PubMed]
- 64.Lee AG (2001) Myelin: delivery by raft. Curr Biol 11:R60–R62 [DOI] [PubMed]
- 65.Kramer EM, Klein C, Koch T, Boytinck M, Trotter J (1999) Compartmentation of Fyn kinase with glycosylphosphatidylinositol-anchored molecules in oligodendrocytes facilitates kinase activation during myelination. J Biol Chem 274:29042–29049 [DOI] [PubMed]
- 66.Klein C, Kramer EM, Cardine AM, Schraven B, Brandt R, Trotter J (2002) Process outgrowth of oligodendrocytes is promoted by interaction of fyn kinase with the cytoskeletal protein tau. J Neurosci 22:698–707 [DOI] [PMC free article] [PubMed]
- 67.Colognato H, Ramachandrappa S, Olsen IM, Ffrench-Constant C (2004) Integrins direct Src family kinases to regulate distinct phases of oligodendrocyte development. J Cell Biol 167:365–375 [DOI] [PMC free article] [PubMed]
- 68.Liang X, Draghi NA, Resh MD (2004) Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J Neurosci 24:7140–7149 [DOI] [PMC free article] [PubMed]
- 69.Stoss O, Novoyatleva T, Gencheva M, Olbrich M, Benderska N, Stamm S (2004) p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol Cell Neurosci 27:8–21 [DOI] [PubMed]
- 70.White R, Gonsior C, Kramer-Albers EM, Stohr N, Huttelmaier S, Trotter J (2008) Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J Cell Biol 181:579–586 [DOI] [PMC free article] [PubMed]
- 71.Brumwell C, Antolik C, Carson JH, Barbarese E (2002) Intracellular trafficking of hnRNP A2 in oligodendrocytes. Exp Cell Res 279:310–320 [DOI] [PubMed]
- 72.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH (1993) Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol 123:431–441 [DOI] [PMC free article] [PubMed]
- 73.Boggs JM (2006) Myelin basic protein: a multifunctional protein. Cell Mol Life Sci 63:1945–1961 [DOI] [PMC free article] [PubMed]
- 74.Musse AA, Gao W, Homchaudhuri L, Boggs JM, Harauz G (2008) Myelin basic protein as a "PI(4, 5) P2-modulin": a new biological function for a major central nervous system protein. Biochemistry 47:10372–10382 [DOI] [PubMed]
- 75.Musse AA, Gao W, Rangaraj G, Boggs JM, Harauz G (2009) Myelin basic protein co-distributes with other PI(4, 5) P(2)-sequestering proteins in Triton X-100 detergent-resistant membrane microdomains. Neurosci Lett 450:32–36 [DOI] [PubMed]
- 76.Nawaz S, Kippert A, Saab A, Werner HB, Lang T, Nave KA, Simons M (2009) Phosphatidylinositol (4,5) bisphosphate regulates membrane targeting of myelin basic protein. J Neurosci 29:4794-4807 [DOI] [PMC free article] [PubMed]
- 77.Chou FC, Chou CH, Shapira R, Kibler RF (1976) Basis of microheterogeneity of myelin basic protein. J Biol Chem 251:2671–2679 [PubMed]
- 78.Kim JK, Mastronardi FG, Wood DD, Lubman DM, Zand R, Moscarello MA (2003) Multiple sclerosis: an important role for post-translational modifications of myelin basic protein in pathogenesis. Mol Cell Proteomics 2:453–462 [DOI] [PubMed]
- 79.Roach A, Takahashi N, Pravtcheva D, Ruddle F, Hood L (1985) Chromosomal mapping of mouse myelin basic protein gene and structure and transcription of the partially deleted gene in shiverer mutant mice. Cell 42:149–155 [DOI] [PubMed]
- 80.O′Connor LT, Goetz BD, Kwiecien JM, Delaney KH, Fletch AL, Duncan ID (1999) Insertion of a retrotransposon in Mbp disrupts mRNA splicing and myelination in a new mutant rat. J Neurosci 19:3404–3413 [DOI] [PMC free article] [PubMed]
- 81.Biffiger K, Bartsch S, Montag D, Aguzzi A, Schachner M, Bartsch U (2000) Severe hypomyelination of the murine CNS in the absence of myelin-associated glycoprotein and fyn tyrosine kinase. J Neurosci 20:7430–7437 [DOI] [PMC free article] [PubMed]
- 82.Sperber BR, Boyle-Walsh EA, Engleka MJ, Gadue P, Peterson AC, Stein PL, Scherer SS, McMorris FA (2001) A unique role for Fyn in CNS myelination. J Neurosci 21:2039–2047 [DOI] [PMC free article] [PubMed]
- 83.Lu Z, Ku L, Chen Y, Feng Y (2005) Developmental abnormalities of myelin basic protein expression in fyn knock-out brain reveal a role of Fyn in posttranscriptional regulation. J Biol Chem 280:389–395 [DOI] [PubMed]
- 84.Morris SJ, Louis CF, Shooter EM (1971) Separation of myelin proteins on two different polyacrylamide gel systems. Neurobiology 1:64–67
- 85.Morell P, Greenfield S, Costantino-Ceccarini E, Wisniewski H (1972) Changes in the protein composition of mouse brain myelin during development. J Neurochem 19:2545–2554 [DOI] [PubMed]
- 86.Magno-Sumbilla C, Campagnoni AT (1977) Factors affecting the electrophoretic analysis of myelin proteins: application to changes occurring during brain development. Brain Res 126:131–148 [DOI] [PubMed]
- 87.Banik NL, Smith ME (1977) Protein determinants of myelination in different regions of developing rat central nervous system. Biochem J 162:247–255 [DOI] [PMC free article] [PubMed]
- 88.Deber CM, Reynolds SJ (1991) Central nervous system myelin: structure, function, and pathology. Clin Biochem 24:113–134 [DOI] [PMC free article] [PubMed]
- 89.Garbay B, Heape AM, Sargueil F, Cassagne C (2000) Myelin synthesis in the peripheral nervous system. Prog Neurobiol 61:267–304 [DOI] [PubMed]
- 90.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R (2006) Molecular anatomy of a trafficking organelle. Cell 127:831–846 [DOI] [PubMed]
- 91.Garbern JY, Cambi F, Tang XM, Sima AA, Vallat JM, Bosch EP, Lewis R, Shy M, Sohi J, Kraft G, Chen KL, Joshi I, Leonard DG, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J (1997) Proteolipid protein is necessary in peripheral as well as central myelin. Neuron 19:205–218 [DOI] [PubMed]
- 92.Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA (2003) Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet 33:366–374 [DOI] [PubMed]
- 93.Yates JR 3rd, Gilchrist A, Howell KE, Bergeron JJ (2005) Proteomics of organelles and large cellular structures. Nat Rev Mol Cell Biol 6:702–714 [DOI] [PubMed]
- 94.Gorg A, Weiss W, Dunn MJ (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4:3665–3685 [DOI] [PubMed]
- 95.Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, Pfeiffer SE (2004) Proteomic mapping provides powerful insights into functional myelin biology. Proc Natl Acad Sci U S A 101:4643–4648 [DOI] [PMC free article] [PubMed]
- 96.Taylor CM, Pfeiffer SE (2003) Enhanced resolution of glycosylphosphatidylinositol-anchored and transmembrane proteins from the lipid-rich myelin membrane by two-dimensional gel electrophoresis. Proteomics 3:1303–1312 [DOI] [PubMed]
- 97.Vanrobaeys F, Van Coster R, Dhondt G, Devreese B, Van Beeumen J (2005) Profiling of myelin proteins by 2D-gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF-TOF mass spectrometry. J Proteome Res 4:2283–2293 [DOI] [PubMed]
- 98.Quarles RH (2007) Myelin-associated glycoprotein (MAG): past, present and beyond. J Neurochem 100:1431–1448 [DOI] [PubMed]
- 99.Johns TG, Bernard CC (1999) The structure and function of myelin oligodendrocyte glycoprotein. J Neurochem 72:1–9 [DOI] [PubMed]
- 100.Birling MC, Tait S, Hardy RJ, Brophy PJ (1999) A novel rat tetraspan protein in cells of the oligodendrocyte lineage. J Neurochem 73:2600–2608 [DOI] [PubMed]
- 101.Werner H, Dimou L, Klugmann M, Pfeiffer S, Nave KA (2001) Multiple splice isoforms of proteolipid M6B in neurons and oligodendrocytes. Mol Cell Neurosci 18:593–605 [DOI] [PubMed]
- 102.Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA (1999) CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99:649–659 [DOI] [PubMed]
- 103.Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S (1999) Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol 145:579–588 [DOI] [PMC free article] [PubMed]
- 104.Devaux J, Gow A (2008) Tight junctions potentiate the insulative properties of small CNS myelinated axons. J Cell Biol 183:909–921 [DOI] [PMC free article] [PubMed]
- 105.Yamaguchi Y, Pfeiffer SE (1999) Highly basic myelin and oligodendrocyte proteins analyzed by NEPHGE-two-dimensional gel electrophoresis: recognition of novel developmentally regulated proteins. J Neurosci Res 56:199–205 [DOI] [PubMed]
- 106.Hartinger J, Stenius K, Hogemann D, Jahn R (1996) 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal Biochem 240:126–133 [DOI] [PubMed]
- 107.Navarre C, Degand H, Bennett KL, Crawford JS, Mortz E, Boutry M (2002) Subproteomics: identification of plasma membrane proteins from the yeast Saccharomyces cerevisiae. Proteomics 2:1706–1714 [DOI] [PubMed]
- 108.Braun RJ, Kinkl N, Beer M, Ueffing M (2007) Two-dimensional electrophoresis of membrane proteins. Anal Bioanal Chem 389:1033–1045 [DOI] [PubMed]
- 109.Yamaguchi Y, Miyagi Y, Baba H (2008) Two-dimensional electrophoresis with cationic detergents: a powerful tool for the proteomic analysis of myelin proteins. Part 2: analytical aspects. J Neurosci Res 86:766–775 [DOI] [PubMed]
- 110.Yamaguchi Y, Miyagi Y, Baba H (2008) Two-dimensional electrophoresis with cationic detergents, a powerful tool for the proteomic analysis of myelin proteins. Part 1: technical aspects of electrophoresis. J Neurosci Res 86:755–765 [DOI] [PubMed]
- 111.Helling S, Schmitt E, Joppich C, Schulenborg T, Mullner S, Felske-Muller S, Wiebringhaus T, Becker G, Linsenmann G, Sitek B, Lutter P, Meyer HE, Marcus K (2006) 2-D differential membrane proteome analysis of scarce protein samples. Proteomics 6:4506–4513 [DOI] [PubMed]
- 112.Fournier ML, Gilmore JM, Martin-Brown SA, Washburn MP (2007) Multidimensional separations-based shotgun proteomics. Chem Rev 107:3654–3686 [DOI] [PubMed]
- 113.Silva JC, Denny R, Dorschel C, Gorenstein MV, Li GZ, Richardson K, Wall D, Geromanos SJ (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol Cell Proteomics 5:589–607 [DOI] [PubMed]
- 114.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5:144–156 [DOI] [PubMed]
- 115.Weinzierl AO, Rudolf D, Hillen N, Tenzer S, van Endert P, Schild H, Rammensee HG, Stevanovic S (2008) Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur J Immunol 38:1503–1510 [DOI] [PubMed]
- 116.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123 [DOI] [PMC free article] [PubMed]
- 117.Tait S, Gunn-Moore F, Collinson JM, Huang J, Lubetzki C, Pedraza L, Sherman DL, Colman DR, Brophy PJ (2000) An oligodendrocyte cell adhesion molecule at the site of assembly of the paranodal axo-glial junction. J Cell Biol 150:657–666 [DOI] [PMC free article] [PubMed]
- 118.Charles P, Tait S, Faivre-Sarrailh C, Barbin G, Gunn-Moore F, Denisenko-Nehrbass N, Guennoc AM, Girault JA, Brophy PJ, Lubetzki C (2002) Neurofascin is a glial receptor for the paranodin/Caspr-contactin axonal complex at the axoglial junction. Curr Biol 12:217–220 [DOI] [PubMed]
- 119.Sherman DL, Tait S, Melrose S, Johnson R, Zonta B, Court FA, Macklin WB, Meek S, Smith AJ, Cottrell DF, Brophy PJ (2005) Neurofascins are required to establish axonal domains for saltatory conduction. Neuron 48:737–742 [DOI] [PubMed]
- 120.Zonta B, Tait S, Melrose S, Anderson H, Harroch S, Higginson J, Sherman DL, Brophy PJ (2008) Glial and neuronal isoforms of Neurofascin have distinct roles in the assembly of nodes of Ranvier in the central nervous system. J Cell Biol 181:1169–1177 [DOI] [PMC free article] [PubMed]
- 121.Spiegel I, Adamsky K, Eisenbach M, Eshed Y, Spiegel A, Mirsky R, Scherer SS, Peles E (2006) Identification of novel cell-adhesion molecules in peripheral nerves using a signal-sequence trap. Neuron Glia Biol 2:27–38 [DOI] [PMC free article] [PubMed]
- 122.Maurel P, Einheber S, Galinska J, Thaker P, Lam I, Rubin MB, Scherer SS, Murakami Y, Gutmann DH, Salzer JL (2007) Nectin-like proteins mediate axon Schwann cell interactions along the internode and are essential for myelination. J Cell Biol 178:861–874 [DOI] [PMC free article] [PubMed]
- 123.Spiegel I, Adamsky K, Eshed Y, Milo R, Sabanay H, Sarig-Nadir O, Horresh I, Scherer SS, Rasband MN, Peles E (2007) A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat Neurosci 10:861–869 [DOI] [PMC free article] [PubMed]
- 124.Park J, Liu B, Chen T, Li H, Hu X, Gao J, Zhu Y, Zhu Q, Qiang B, Yuan J, Peng X, Qiu M (2008) Disruption of Nectin-like 1 cell adhesion molecule leads to delayed axonal myelination in the CNS. J Neurosci 28:12815–12819 [DOI] [PMC free article] [PubMed]
- 125.Stipp CS, Kolesnikova TV, Hemler ME (2001) EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J Biol Chem 276:40545–40554 [DOI] [PubMed]
- 126.Stipp CS, Kolesnikova TV, Hemler ME (2003) EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J Cell Biol 163:1167–1177 [DOI] [PMC free article] [PubMed]
- 127.Terada N, Baracskay K, Kinter M, Melrose S, Brophy PJ, Boucheix C, Bjartmar C, Kidd G, Trapp BD (2002) The tetraspanin protein, CD9, is expressed by progenitor cells committed to oligodendrogenesis and is linked to beta1 integrin, CD81, and Tspan-2. Glia 40:350–359 [DOI] [PubMed]
- 128.Ishibashi T, Ding L, Ikenaka K, Inoue Y, Miyado K, Mekada E, Baba H (2004) Tetraspanin protein CD9 is a novel paranodal component regulating paranodal junctional formation. J Neurosci 24:96–102 [DOI] [PMC free article] [PubMed]
- 129.Ji B, Case LC, Liu K, Shao Z, Lee X, Yang Z, Wang J, Tian T, Shulga-Morskaya S, Scott M, He Z, Relton JK, Mi S (2008) Assessment of functional recovery and axonal sprouting in oligodendrocyte-myelin glycoprotein (OMgp) null mice after spinal cord injury. Mol Cell Neurosci 39:258–267 [DOI] [PMC free article] [PubMed]
- 130.Steen H, Mann M (2004) The ABC′s (and XYZ′s) of peptide sequencing. Nat Rev Mol Cell Biol 5:699–711 [DOI] [PubMed]
- 131.Buser AM, Erne B, Werner HB, Nave KA, Schaeren-Wiemers N (2008) The septin cytoskeleton in myelinating glia. Mol Cell Neurosci 40:156–166 [DOI] [PubMed]
- 132.Brockschnieder D, Sabanay H, Riethmacher D, Peles E (2006) Ermin, a myelinating oligodendrocyte-specific protein that regulates cell morphology. J Neurosci 26:757–762 [DOI] [PMC free article] [PubMed]
- 133.Thurnherr T, Benninger Y, Wu X, Chrostek A, Krause SM, Nave KA, Franklin RJ, Brakebusch C, Suter U, Relvas JB (2006) Cdc42 and Rac1 signaling are both required for and act synergistically in the correct formation of myelin sheaths in the CNS. J Neurosci 26:10110–10119 [DOI] [PMC free article] [PubMed]
- 134.Anitei M, Cowan AE, Pfeiffer SE, Bansal R (2008) Role for Rab3a in oligodendrocyte morphological differentiation. J Neurosci Res 87:342–352 [DOI] [PubMed]
- 135.Schardt A, Brinkmann BG, Mitkovski M, Sereda MW, Werner HB, Nave K-A (2009) The SNARE protein SNAP-29 interacts with the GTPase Rab3A: implications for membrane trafficking in myelinating glia. J Neurosci Res doi:10.1002/jnr.22005 [DOI] [PubMed]
- 136.Golan N, Adamsky K, Kartvelishvily E, Brockschnieder D, Mobius W, Spiegel I, Roth AD, Thomson CE, Rechavi G, Peles E (2008) Identification of Tmem10/Opalin as an oligodendrocyte enriched gene using expression profiling combined with genetic cell ablation. Glia 56:1176–1186 [DOI] [PMC free article] [PubMed]
- 137.Kippert A, Trajkovic K, Fitzner D, Opitz L, Simons M (2008) Identification of Tmem10/Opalin as a novel marker for oligodendrocytes using gene expression profiling. BMC Neurosci 9:40 [DOI] [PMC free article] [PubMed]
- 138.Yoshikawa F, Sato Y, Tohyama K, Akagi T, Hashikawa T, Nagakura-Takagi Y, Sekine Y, Morita N, Baba H, Suzuki Y, Sugano S, Sato A, Furuichi T (2008) Opalin, a transmembrane sialylglycoprotein located in the central nervous system myelin paranodal loop membrane. J Biol Chem 283:20830–20840 [DOI] [PMC free article] [PubMed]
- 139.Morris JK, Willard BB, Yin X, Jeserich G, Kinter M, Trapp BD (2004) The 36K protein of zebrafish CNS myelin is a short-chain dehydrogenase. Glia 45:378–391 [DOI] [PubMed]
- 140.Garbern JY (2007) Pelizaeus–Merzbacher disease: genetic and cellular pathogenesis. Cell Mol Life Sci 64:50–65 [DOI] [PMC free article] [PubMed]
- 141.Swatton JE, Prabakaran S, Karp NA, Lilley KS, Bahn S (2004) Protein profiling of human postmortem brain using 2-dimensional fluorescence difference gel electrophoresis (2-D DIGE). Mol Psychiatry 9:128–143 [DOI] [PubMed]
- 142.Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A (2007) Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochem Res 32:187–195 [DOI] [PubMed]
- 143.Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, Guo J, Ling EA, Liang F (2007) Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci 27:2606–2616 [DOI] [PMC free article] [PubMed]
- 144.Rasband MN, Tayler J, Kaga Y, Yang Y, Lappe-Siefke C, Nave KA, Bansal R (2005) CNP is required for maintenance of axon-glia interactions at nodes of Ranvier in the CNS. Glia 50:86–90 [DOI] [PubMed]
- 145.Bifulco M, Laezza C, Stingo S, Wolff J (2002) 2′, 3′-Cyclic nucleotide 3′-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc Natl Acad Sci U S A 99:1807–1812 [DOI] [PMC free article] [PubMed]
- 146.Lee J, Gravel M, Zhang R, Thibault P, Braun PE (2005) Process outgrowth in oligodendrocytes is mediated by CNP, a novel microtubule assembly myelin protein. J Cell Biol 170:661–673 [DOI] [PMC free article] [PubMed]
- 147.Polevoda B, Sherman F (2002) The diversity of acetylated proteins. Genome Biol 3(5):reviews0006 [DOI] [PMC free article] [PubMed]
- 148.Shimazu T, Horinouchi S, Yoshida M (2007) Multiple histone deacetylases and the CREB-binding protein regulate pre-mRNA 3′-end processing. J Biol Chem 282:4470–4478 [DOI] [PubMed]
- 149.Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY, Yeo CY, Lee KY (2008) Sirt2 interacts with 14–3-3 beta/gamma and down-regulates the activity of p53. Biochem Biophys Res Commun 368:690–695 [DOI] [PubMed]
- 150.Flores AI, Narayanan SP, Morse EN, Shick HE, Yin X, Kidd G, Avila RL, Kirschner DA, Macklin WB (2008) Constitutively active Akt induces enhanced myelination in the CNS. J Neurosci 28:7174–7183 [DOI] [PMC free article] [PubMed]
- 151.Mrowiec T, Schwappach B (2006) 14–3-3 proteins in membrane protein transport. Biol Chem 387:1227–1236 [DOI] [PubMed]
- 152.Berdichevsky A, Guarente L (2006) A stress response pathway involving sirtuins, forkheads and 14-3-3 proteins. Cell Cycle 5:2588–2591 [DOI] [PubMed]
- 153.Hurd TW, Fan S, Liu CJ, Kweon HK, Hakansson K, Margolis B (2003) Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol 13:2082–2090 [DOI] [PubMed]
- 154.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B (2003) Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol 5:137–142 [DOI] [PubMed]
- 155.Chan JR, Jolicoeur C, Yamauchi J, Elliott J, Fawcett JP, Ng BK, Cayouette M (2006) The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science 314:832–836 [DOI] [PubMed]
- 156.Han MH, Hwang SI, Roy DB, Lundgren DH, Price JV, Ousman SS, Fernald GH, Gerlitz B, Robinson WH, Baranzini SE, Grinnell BW, Raine CS, Sobel RA, Han DK, Steinman L (2008) Proteomic analysis of active multiple sclerosis lesions reveals therapeutic targets. Nature 451:1076–1081 [DOI] [PubMed]