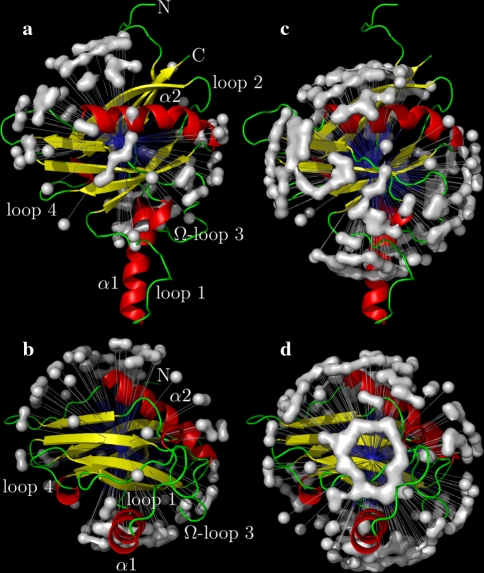
Fig. 4.
The OMP backbone amide NH bond vector orientations employed in (a, b) the original model-free analysis of Gitti et al. (2005) and (c, d) in the reanalysis using the new model-free optimisation protocol. In the original analysis the diffusion tensor was determined using residues solely within the strands of the β clam fold and helix α2 whereas in the reanalysis all residues were used. The distributions correspond surfaces draped over artificial NH vectors with the nitrogen positioned at the centre of mass of all selected residues and the bond length being set to 20 Å. Because of the symmetry of spheroidal and ellipsoidal diffusion tensors the positive or negative orientation of the XH bond has no effect on relaxation and, hence, a second artificial NH vector has been added for each residue whereby the orientation has been reversed. The PyMOL images were generated using relax