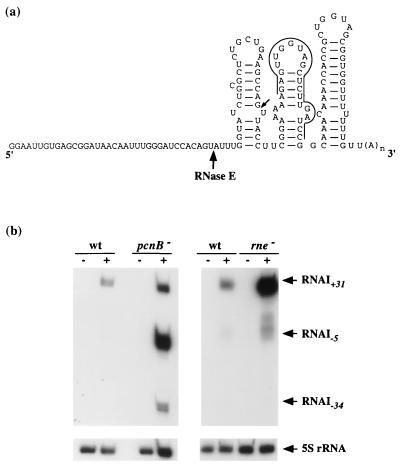
Figure 3.
(a) Schematic structure of the engineered RNAI (RNAI+31) from ENS304 cells. The three hairpins are tentatively drawn based on the structure of the genuine RNAI. The positions of the major and minor RNase E cleavage sites, generating RNAI−5 and RNAI−34, respectively, are indicated by an arrow. The 5′ extension corresponds mostly to the sequence transcribed from the lac operator: the genuine RNAI sequence starts 5 nt upstream of the major RNase E site. The sequence complementary to the oligonucleotide probe used for Northern blots and primer extensions is overlined. (b) Northern blots showing the RNAI+31, RNAI−5, and RNAI−34 species (arrow) in the wild-type ENS304 strain and pcnB80 derivative (Left) or in the wild-type and rne-1 derivative (Right). (Left) Cells were grown steadily at 37°C. (Right) Cells were grown at 30°C and then shifted to 42°C 30 min before harvest. Signal intensities in both are comparable directly. The symbols (+) and (−) denote the presence or absence of IPTG in the growth medium. In the bottom panels, the same membrane has been reprobed with a 5S rRNA-specific probe.