Abstract
Amelogenin and ameloblastin, the major enamel matrix proteins, are important for enamel mineralization. To identify their synergistic roles in enamel development, we generated Amel X-/-/Ambn-/- mice. These mice showed additional enamel defects in comparison with Amel X-/- or Ambn-/- mice. In 7-day-old Amel X-/-/Ambn-/- mice, not only was the ameloblast layer irregular and detached from the enamel surface, as in Ambn-/-, but also, the enamel width was significantly reduced in the double-null mice as compared with Amel X-/- or Ambn-/- mice. Proteomic analysis of the double-null teeth revealed increased levels of RhoGDI (Arhgdia), a Rho-family-specific guanine nucleotide dissociation inhibitor, which is involved in important cellular processes, such as cell attachment. Both Amel X-/-/Ambn-/- mice and Ambn-/- mice displayed positive staining with RhoGDI antibody in the irregularly shaped ameloblasts detached from the matrix. Ameloblastin-regulated expression of RhoGDI suggests that Rho-mediated signaling pathway might play a role in enamel formation.
Keywords: enamel, amelogenin, ameloblastin, knockout mice, RhoGDI (Arhgdia)
Introduction
Dental enamel is the most highly mineralized tissue in the body, and is formed as a result of mineralization of enamel matrices secreted by ameloblasts. Ameloblasts secrete several enamel matrix proteins, such as amelogenins, ameloblastin, and enamelin. These enamel matrix proteins are processed and degraded by proteases such as MMP20 and KLK4 during enamel mineralization (Bartlett et al., 1996; Simmer et al., 1998). The highly orchestrated secretion of enamel matrix proteins and their proper degradation are critical for normal enamel formation.
The amelogenin proteins are highly conserved across species, and constitute 90% of the enamel organic matrix. Based on the results from our study of Amel X-/- mice, amelogenins play an important role in enamel biomineralization (Gibson et al., 2001; Hatakeyama et al., 2003). In the Amel X-/- mice, ameloblast differentiation was relatively normal, but an abnormally thin enamel layer was formed (Gibson et al., 2001, 2005).
It was concluded that amelogenins are essential for well-organized hydroxyapatite prism formation and elongation during enamel development, and for producing normal enamel thickness, but not for the initiation of enamel formation. Our recent studies on transgenic mice, which express the most abundant amelogenin form, M180, in the amelogenin null background, demonstrated that M180 could significantly rescue the enamel defects of the amelogenin null mice (Li et al., 2008). Self-assembly of amelogenin proteins into nanospheres has been recognized as a key factor in controlling the orientation and elongated growth of crystals during the mineralizing process in enamel (Du et al., 2005). Transgenic mice that express an amelogenin protein with a mutation either at the N or C terminus showed that the N-terminal domain of amelogenin might be involved in the formation of nanospheres (Paine et al., 2003a), whereas the C-terminal region could contribute to stability and homogeneity in sizes of nanospheres, preventing mineral crystal fusion to form larger structures prematurely (Moradian-Oldak and Goldberg, 2005; Moradian-Oldak et al., 2006). In addition, we have recently reported amelogenin’s function for osteoclast differentiation in periodontal ligament tissue (Hatakeyama et al., 2006).
Ameloblastin, also known as amelin or sheathlin, is an enamel-specific glycoprotein, which is the most abundant non-amelogenin enamel matrix protein (Cerný et al., 1996; Krebsbach et al., 1996; Fong et al., 1998), and serves as a cell adhesion molecule for ameloblasts, but not for dental epithelial cells (Fukumoto et al., 2004, 2005). Ameloblastin expression in ameloblasts peaks at the secretory stage and diminishes at the maturation stage. Transgenic mice overexpressing ameloblastin in ameloblasts have impaired enamel structures, suggesting the importance of normal levels of ameloblastin in enamel formation (Paine et al., 2003b). Furthermore, in Ambn-/- mice, the dental epithelium differentiates into enamel-secreting ameloblasts, but the cells detach from the matrix surface at the secretory stage and lose polarity. In ameloblasts of Ambn-/- teeth, the expression of amelogenins is reduced to about 20% of that of Ambn+/+ teeth, while other enamel matrix proteins are expressed at nearly normal levels (Fukumoto et al., 2004). These results suggested that ameloblastin is essential in maintaining normal ameloblast differentiation and attachment to the enamel matrix. Thus, the cellular functions of amelogenin and ameloblastin are apparently distinct, and in this paper we report potential synergistic functions of these 2 enamel proteins.
Materials & Methods
Amel X-/-/Ambn-/- Mice
Targeted disruption of amelogenin (Amel X) and ameloblastin (Ambn) genes has been described previously (Gibson et al., 2001; Fukumoto et al., 2004). Amel X-/- mice were mated with Ambn-/- mice to generate double-heterozygous mice, which were interbred to generate Amel X-/-/Ambn-/- mice. (Detailed information on generation and genotyping is described in the online Appendix and Appendix Fig. 1.) Mutant mice were initially analyzed in a C57BL/6 × 129/SvEv mixed genetic background and later in an enriched C57BL/6 background by being back-crossed 4 x with C57BL/6 mice. Standard NIH guidelines were followed for housing, feeding, and breeding the mice. These studies were carried out with the approval of the NIDCR Animal Care and Use Committee.
Scanning Electron Microscopic (SEM) Analyses of Incisors and Molars
Incisors and molars from wild-type and mutant mice were coated with gold and photographed by scanning electron microscopy at 20 kV (Jeol JSM T330A, Jeol, Inc., Peabody, MA, USA), and energy-dispersive spectroscopy (Kevex X-ray, Scotts Valley, CA, USA).
To observe the enamel crystals, we embedded the specimens in epoxy resin, cut them with an ISOMET low-speed saw (Buehler, Lake Bluff, IL, USA), treated them with 40% phosphoric acid for 10 sec and 10% sodium hypochlorite for 30 sec, and then coated them with gold.
Preparation of Tissue Sections and Immunohistochemistry
Post-natal (P) days 1 (P1) and 7 (P7) mouse skulls were dissected and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 16 hrs at 4°C. Tissues were decalcified with 250 mM EDTA/PBS and embedded in paraffin for paraffin sections or in OCT compound (Sakura Finetechnical Co., Torrance, CA, USA) for frozen sectioning. Frozen sections were cut at 8-µm intervals on a cryostat (2800 Frigocut, Leica Inc., Wetzlar, Germany). Paraffin sections were cut at 5-µm intervals on a microtome (RM2155, Leica Inc.). For detailed morphological analysis,sections were stained with hematoxylin and eosin Y (Sigma, St. Louis, MO, USA). Frozen sections were immunostained for RhoGDI with goat polyclonal antibodies against mouse RhoGDI (Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C at a dilution of 1:100. After being washed with PBS, the sections were incubated with peroxidase-conjugated mouse antibodies against goat IgG (Vector Laboratories, Burlingame, CA, USA), treated with diaminobenzidine substrate, and counterstained with hematoxylin for light microscopy. For control, frozen sections were incubated with secondary antibody only.
Results
Defective Enamel Formation in Amel X-/-/Ambn-/- Mice
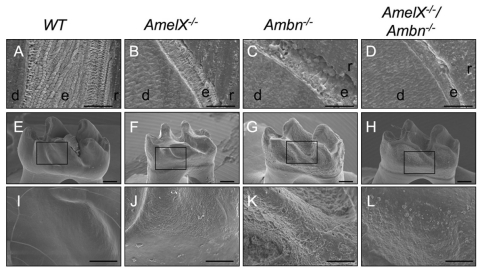
SEM analysis of incisors revealed hypoplastic enamel and a lack of prism pattern in Amel X-/-, Ambn-/-, and Amel X-/-/Ambn-/- mice that is the hallmark of organized mineral crystals in normal enamel (Figs. 1A-1D). Enamel width was much thinner in Amel X-/-/Ambn-/- mice as compared with that in Amel X-/- and Ambn-/- mice. As in Ambn-/- mice, flat plate-like structures extended perpendicularly from the enamel surface to the dentin enamel junction in Amel X-/-/Ambn-/- mice (Figs. 1C, 1D). The enamel surfaces appeared cobbled in both Amel X-/- (Fig. 1J) and Ambn-/- (Fig. 1K) mice. However, in Amel X-/-/Ambn-/- mice, the molar surfaces appeared less cobbled than in Amel X-/- and Ambn-/- mice (Fig. 1L). Elemental analysis indicated that the composition was similar to that of hydroxyapatite, indicating a normal formation of mineral in the absence of the amelogenin and ameloblastin proteins. The Ca/P molar ratio was also not significantly different in the teeth of all null mice and the WT controls (almost 1.5; data not shown).
Figure 1.
Scanning electron microscopy analysis of teeth from Amel X-/-, Ambn-/-, Amel X-/-/Ambn-/-, and wild-type mice. (A-D) Incisors from the 6-week-old mutant and wild-type mice; the enamel (e) in junction with dentin (d) is shown. Note the thin aprismatic structure in Amel X-/- mice (B). The enamel width of Amel X-/-/Ambn-/- (D) mice markedly reduced as compared with that of the Ambn-/- mice (C). (E-L) Molars of the 6-week-old wild-type and mutant mice; note the small crown size of Amel X-/- mice (F) and the double-mutant (H). The enamel from Amel X-/-(F), Ambn-/- (G), and Amel X-/-/Ambn-/- (H) appeared abnormal as compared with that in the wild-type mice (E). (I-L) Teeth from all 3 mutant mice mimic the amelogenesis imperfecta phenotype. Amel X-/-/Ambn-/- enamel appeared less cobbled as compared with Ambn-/- enamel. Bars in A-D = 50 µm; bars in E-L = 250 µm. e, enamel; d, dentin; r, resin.
Unlike Ambn-/- Ameloblasts, Amel X-/-/Ambn-/- Ameloblasts Do Not Develop Calcified Nodules
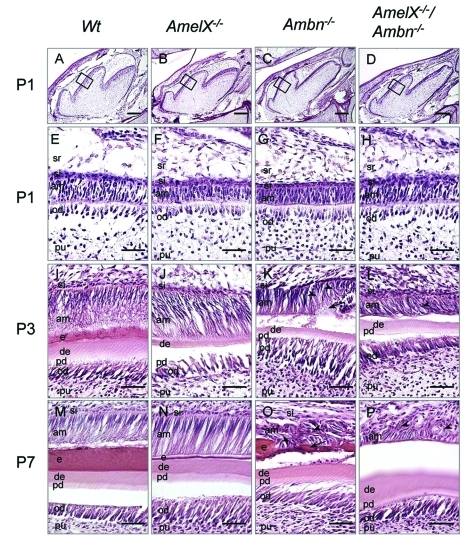
In early stages of molar development up to P1, no differences were observed in either shape or size of the tooth buds of WT, Amel X-/-, Ambn-/-, and Amel X-/-/Ambn-/- mice (Figs. 2A-2D). At P1, dentin formation of molars had begun, and dental epithelium had started to elongate and polarize with the apical nuclear localization in all of these mice (Figs. 2E-2H). Thus, cellular organization of ameloblasts and odontoblasts was similar in these mice at the pre-secretory stage. However, at P3, ameloblasts of Ambn–/– and Amel X-/-/Ambn-/- mice started to detach from the matrix layer and lost the cell polarity with the centralized nuclear localization (Figs. 2K, 2L), whereas normal ameloblasts were polarized, elongated, and formed an enamel matrix in WT and Amel X-/- mice (Figs. 2I, 2J). At P7, Ambn–/– and Amel X-/-/Ambn-/- ameloblasts completely lost their polarity (short and round) and accumulated to form a multilayered structure (Figs. 2O, 2P, arrowhead), in contrast to the single layer of WT and Amel X-/- ameloblasts (Figs. 2M, 2N). Interestingly, Ambn-/- ameloblasts contained calcified nodules (Fig. 2O, arrow), but Amel X-/-/Ambn-/- cells did not (Fig. 2P).
Figure 2.
Histological analysis of teeth from the wild-type, Amel X-/-, Ambn-/-, and Amel X-/-/Ambn-/- mice. Hematoxylin-eosin staining of the sagittal sections of the mandibular first molars of P1 (A-H), P3 (I-L), and P7 (M-P) wild-type and mutant mice: wild-type (A,E,I,M), Amel X-/- (B,F,J,N), Ambn-/- (C,G,K,O), and Amel X-/-/Ambn-/- mice (D,H,L,P). P3 and P7 Ambn–/– ameloblasts display multiple layers containing abnormal calcified structures (Figs. 2K and 2O, arrows). Amel X-/-/Ambn-/- ameloblasts also form multiple layers; however, they do not contain the calcified structures (Figs. 2L and 2P, arrowhead). am, ameloblast; si, stratum intermedium; e, enamel; pd, predentin; de, dentin; od, odontoblast; pu, pulp; sr, stellate reticulum. Bars in A-D = 500 µm; bars in E-P = 50 µm.
Increased RhoGDI Expression in Amel X-/-/Ambn-/- Ameloblasts
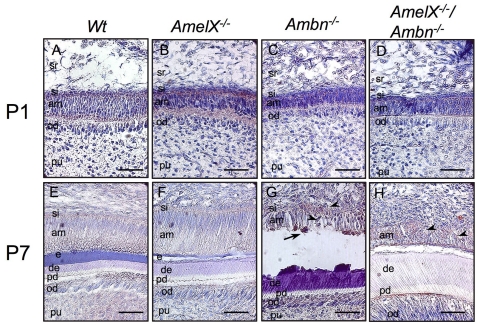
We utilized proteomic analysis to identify 24-kDa-size protein, which was increased in Amel X-/-/Ambn-/- ameloblasts (Appendix Fig. 2). Using MALDI analysis, we identified this protein as RhoGDI. For further analysis of RhoGDI expression patterns in developing mouse molars, we carried out immunohistochemical analysis. At P1, weak RhoGDI expression was observed in ameloblasts and odontoblasts of the WT, Amel X-/-, Ambn-/-, and Amel X-/-/Ambn-/- mice (Figs. 3A-3D). However, at P7, the ameloblasts of WT and Amel X-/- mice had no noticeable RhoGDI expression (Figs. 3E, 3F), whereas irregularly shaped ameloblasts in Ambn-/- and Amel X-/-/Ambn-/- mice showed sustained expression of RhoGDI (Figs. 3G, 3H). Calcified nodules were also detected adjacent to the irregular ameloblast layer in Ambn-/- mice (Fig. 3G, arrow), but not in Amel X-/-/Ambn-/- mice. We also noted increased expression of RhoGDI in the lower first molars of the 7-day-old Amel X-/-/Ambn-/- mice by RT-PCR (Appendix Fig. 3).
Figure 3.
RhoGDI expression in the ameloblasts of the wild-type, Amel X-/-, Ambn-/-, and Amel X-/-/Ambn-/- mice. Sagittal sections of the incisors from the wild-type (A,E), Amel X-/- (B,F), Ambn-/- (C,G), and Amel X-/-/Ambn-/- mice (D,H) were stained with the RhoGDI antibody as described in Materials & Methods. Note positive staining in the detached ameloblasts of Ambn-/- (C,G) and Amel X-/-/Ambn-/- mice (D,H). Bars = 50 µm. am, ameloblast; od, odontoblast; pu, pulp; si, stratum intermedium; e, enamel; d, dentin; pd, predentin.
Discussion
To delineate potential synergistic roles of amelogenins and ameloblastin, we analyzed teeth from the wild-type, Amel X-/-, Ambn-/-, and Amel X-/-/Ambn-/- mice. Our analysis revealed that the Amel X-/-/Ambn-/- mice displayed additional enamel defects. As compared with the Amel X-/- and Ambn-/- mice, enamel width was markedly reduced in Amel X-/-/ Ambn-/- mice. Although ameloblast morphology was similar in Ambn-/- and Amel X-/-/ Ambn-/- mice, calcified nodules observed in Ambn-/- ameloblasts were absent in the double-null ameloblasts. These additional defects in Amel X-/-/Ambn-/- ameloblasts suggest a possible synergism in the cellular functions of amelogenins and ameloblastin.
Surprisingly, Amel X-/-/Ambn-/- mice still showed a very thin layer of enamel, in spite of the lack of the 2 most abundant ECM proteins secreted by ameloblasts to form normal enamel. We found that enamelin was still expressed in the Amel X-/-/Ambn-/- teeth (based on our RT-PCR analysis; data not shown). The enamelin gene (ENAM) has also been implicated in human amelogenesis imperfecta (Kim et al., 2005). Enam-/- mice did not form normal enamel, because of the lack of mineralization at the secretory surfaces of the ameloblasts (Hu et al., 2008). In addition, ENAM point mutation resulted in the phenotype resembling amelogenesis imperfecta (Masuya et al., 2005). Therefore, it is possible that enamelin might be involved in enamel formation in the Amel X-/-/Ambn-/- mice. In addition to enamelin, other ECM proteins might play a role in enamel formation in these mice, and their identification will require further studies. We had previously reported that amelogenins are involved in osteoclast differentiation in PDL cells, and furthermore, one can speculate that its lack in the double-null mice might contribute in some way to the formation of thinner enamel. Interestingly, SEM analysis of molars and incisors indicated smoother enamel in the Amel X-/-/Ambn-/- mice as compared with Ambn-/- enamel. This phenotypic difference can be possibly attributed to the presence of irregular calcified nodules in Ambn-/- ameloblasts, and one can speculate that these nodules are formed because of the residual amelogenin in these mice (Fukumoto et al., 2004).
Our proteomic studies identified an increased protein level of RhoGDI (Arhgdia) in Amel X-/-/Ambn-/- teeth. RhoGDI, a Rho-family-specific guanine nucleotide dissociation inhibitor, forms a tight complex with Rho GTPases and inactivates Rho GTPases function as a cytosolic molecule. Reduced expression or inactivation of RhoGDIs releases Rho GTPases from the complex and translocates Rho GTPases into the membrane for activation of Rho signaling pathways (Takai et al., 1995). Rho GTPases such as Rho, Rac, and Cdc42 are known to regulate assembly of filamentous actin (F-actin) and the organization of the actin cytoskeleton, and the regulation of gene transcription, cell cycle, microtubule dynamics, vesicle transport, and numerous enzymatic activities. In the wild-type teeth, RhoGDI was expressed in undifferentiated dental epithelium, but its expression was down-regulated in the secretory stage of ameloblasts. During tooth development, protein expression of RhoGDI is not altered at the early stage in Amel X-/-, Ambn-/- , and Amel X-/-/Ambn-/- ameloblasts. However, in later stages, when cells continue to proliferate and form multicellular layers in Amel X-/-/Ambn-/- mice, RhoGDI expression is increased. The Rho signaling pathways in murine ameloblasts are known to induce F-actin product (Li et al., 2005). F-actin-rich regions have been described, and these include Tomes’ process, distal terminal webs, and distal ends of ruffled or smooth-ended ameloblasts in rat incisors (Nishikawa and Kitamura, 1986).
Interestingly, the human Amel X gene has been shown to reside in a “nested” gene structure within intron 1 of the ARHGAP6 gene that encodes Rho GAP, which regulates RhoA activity (Hall and Nobes, 2000; Prakash et al., 2005). In some cases, expression of nested and host genes is simultaneously up- and down-regulated by common regulatory elements (Peters and Ross, 2001). It is possible that the expression of Amel X and ARHGAP6 genes might be similarly regulated. Rho is recognized as a molecular switch (Hall and Nobes, 2000), which normally cycles from the active GTP-bound form to the inactive GDP-bound form (Li et al., 2005), thereby regulating downstream events leading to changes in the cytoskeleton. It has been shown that Rac1 and Cdc42, downstream of Rho signaling, are regulators of cell spreading and formation of lamellipodia and filopodia (Clark et al., 1998; Hall, 1998), and cell polarization (Etienne-Manneville and Hall, 2002; Cau and Hall, 2005). Rac1 and Cdc42 regulate laminin-10/11-mediated cell spreading and filopodia formation of the dental epithelium (Fukumoto et al., 2006). Increased expression of RhoGDI in Amel X-/-/Ambn-/- teeth might inhibit active Rho GTP, resulting in irregular ameloblast morphology.
In summary, our study suggests that the enamel matrix proteins such as amelogenins and ameloblastin are not only required for the formation of a proper matrix for well-orchestrated enamel biomineralization, but also have synergistic cellular functions during enamel development.
Supplementary Material
Acknowledgments
We thank Drs. Aya Yamada and Yoko Kamasaki for generous help with SEM analysis, and Harry Grant for editorial assistance.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
This work was supported by the Division of Intramural Research of the National Institute of Dental and Craniofacial Research.
References
- Bartlett JD, Simmer JP, Xue J, Margolis HC, Moreno EC. (1996). Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 183:123-128 [DOI] [PubMed] [Google Scholar]
- Cau J, Hall A. (2005). Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J Cell Sci 118(Pt 12):2579-2587 [DOI] [PubMed] [Google Scholar]
- Cerný R, Slaby I, Hammarström L, Wurtz T. (1996). A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J Bone Miner Res 11:883-891 [DOI] [PubMed] [Google Scholar]
- Clark EA, King WG, Brugge JS, Symons M, Hynes RO. (1998). Integrin-mediated signals regulated by members of the rho family of GTPases. J Cell Biol 142:573-586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. (2005). Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science 307:1450-1454; erratum in Science 309:2166, 2005 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. (2002). Rho GTPases in cell biology. Nature 420:629-635 [DOI] [PubMed] [Google Scholar]
- Fong CD, Cerný R, Hammarström L, Slaby I. (1998). Sequential expression of an amelin gene in mesenchymal and epithelial cells during odontogenesis in rats. Eur J Oral Sci 106(Suppl 1):324-330 [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, et al. (2004). Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol 167:973-983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S, Yamada A, Nonaka K, Yamada Y. (2005). Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation. Cells Tissues Organs 181:189-195 [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Miner JH, Ida H, Fukumoto E, Yuasa K, Miyazaki H, et al. (2006). Laminin alpha5 is required for dental epithelium growth and polarity and the development of tooth bud and shape. J Biol Chem 281:5008-5016 [DOI] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, et al. (2001). Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem 276:31871-31875 [DOI] [PubMed] [Google Scholar]
- Gibson CW, Kulkarni AB, Wright JT. (2005). The use of animal models to explore amelogenin variants in amelogenesis imperfecta. Cells Tissues Organs 181:196-201 [DOI] [PubMed] [Google Scholar]
- Hall A. (1998). Rho GTPases and the actin cytoskeleton. Science 279:509-514 [DOI] [PubMed] [Google Scholar]
- Hall A, Nobes CD. (2000). Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos Trans R Soc Lond B Biol Sci 355:965-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Sreenath T, Hatakeyama Y, Thyagarajan T, Shum L, Gibson CW, et al. (2003). The receptor activator of nuclear factor-kappa B ligand-mediated osteoclastogenic pathway is elevated in amelogenin-null mice. J Biol Chem 278:35743-35748 [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Philp D, Hatakeyama Y, Haruyama N, Shum L, Aragon MA, et al. (2006). Amelogenin-mediated regulation of osteoclastogenesis and periodontal cell proliferation and igration. J Dent Res 85:144-149 [DOI] [PubMed] [Google Scholar]
- Hu JCC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, et al. (2008). Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J Biol Chem 283:10858-10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, et al. (2005). ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res 84:278-282 [DOI] [PubMed] [Google Scholar]
- Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, Yamada Y. (1996). Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J Biol Chem 271:4431-4435 [DOI] [PubMed] [Google Scholar]
- Li Y, Decker S, Yuan ZA, DenBesten PK, Aragon MA, Jordan-Sciutto K, et al. (2005). Effects of sodium fluoride on the actin cytoskeleton of murine ameloblasts. Arch Oral Biol 50:681-688 [DOI] [PubMed] [Google Scholar]
- Li Y, Suggs C, Wright JT, Yuan Z, Aragon M, Fong H, et al. (2008). Partial rescue of the amelogenin null dental enamel phenotype. J Biol Chem 283:15056-15062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, Shimizu A, et al. (2005). Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI). Hum Mol Genet 14:575-583 [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Goldberg M. (2005). Amelogenin supra-molecular assembly in vitro compared with the architecture of the forming enamel matrix. Cells Tissues Organs 181:202-218 [DOI] [PubMed] [Google Scholar]
- Moradian-Oldak J, Du C, Falini G. (2006). On the formation of amelogenin microribbons. Eur J Oral Sci 114(Suppl 1):289-296 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Kitamura H. (1986). Localization of actin during differentiation of the ameloblast, its related epithelial cells and odontoblasts in the rat incisor using NBD-phallacidin. Differentiation 30:237-243 [DOI] [PubMed] [Google Scholar]
- Paine ML, Luo W, Zhu DH, Bringas P, Jr, Snead ML. (2003a). Functional domains for amelogenin revealed by compound genetic defects. J Bone Miner Res 18:466-472 [DOI] [PubMed] [Google Scholar]
- Paine ML, Wang HJ, Luo W, Krebsbach PH, Snead ML. (2003b). A transgenic animal model resembling amelogenesis imperfecta related to ameloblastin overexpression. J Biol Chem 278:19447-19452 [DOI] [PubMed] [Google Scholar]
- Peters MF, Ross CA. (2001). Isolation of a 40-kDa Huntingtin-associated protein. J Biol Chem 276:3188-3194 [DOI] [PubMed] [Google Scholar]
- Prakash SK, Gibson CW, Wright JT, Boyd C, Cormier T, Sierra R, et al. (2005). Tooth enamel defects in mice with a deletion at the Arhgap 6/Amel X locus. Calcif Tissue Int 77:23-29 [DOI] [PubMed] [Google Scholar]
- Simmer JP, Fukae M, Tanabe T, Yamakoshi Y, Uchida T, Xue J, et al. (1998). Purification, characterization, and cloning of enamel matrix serine proteinase 1. J Dent Res 77:377-386 [DOI] [PubMed] [Google Scholar]
- Takai Y, Sasaki T, Tanaka K, Nakanishi H. (1995). Rho as a regulator of the cytoskeleton. Trends Biochem Sci 20:227-231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.