Abstract
The establishment of mouse embryonic stem (ES) cell liness has allowed for the generation of the knockout mouse. ES cells that are genetically altered in culture can then be manipulated to derive a whole mouse containing the desired mutation. To successfully generate a knockout mouse, however, the ES cells must be carefully cultivated in a pluripotent state throughout the gene targeting experiment. This unit describes detailed step-by-step protocols, reagents, equipment, and strategies needed for the successful generation of gene knockout embryonic stem cells using homologous recombination technologies.
Key Terms: mouse embryonic stem cells, homologous recombination
Introduction
The isolation of murine embryonic stem cells (ES) has been an important discovery necessary for the development of mice with designer mutations. Stem cells are capable of self-renewal in culture and can be maintained in an undifferentiated state under certain growth conditions. Stem cells are also totipotent and, when injected into a host blastocyst, can contribute to the somatic and germ cell lineages of the resulting chimeric mouse. If germline transmission occurs, an offspring of the chimeric mouse can be produced that was derived from the injected ES cell clone. The ability to pass on germline transmission means that a mouse can be generated from ES cells that are genetically manipulated in culture. With the discovery of homologous recombination, stem cells were seen as an ideal tool that could be used to make genetically altered mice. Homologous recombination in stem cells was first applied for the development of knockout mice through targeted gene inactivation. Eventually, these techniques were adapted for creating conditional knockout mice, knock-in mice, and mice with subtle mutations such as genetic point mutations, deletions, and insertions. With gene targeting in mice, scientists have been able to create animal models that mimic human genetic diseases. Additionally, these animal models have also provided an important tool for delineating the function of a gene in vivo.
The properties of ES cells include pluripotency and self-renewal in tissue culture. Embryonic stem cells were derived from the inner cell mass of a 3.5-day-old mouse embryo and were grown successfully in vitro (Evans and Kaufman 1981; Martin 1981). These isolated stem cells remain karyotypically normal in culture and do not transform into teratocarcinomas. Under specified growth conditions, ES cells can also be induced to form multiple cell types and even embryoid bodies. This pluripotent property allows scientists to test which growth factors are involved in turning an undifferentiated cell into specified cell types. With the capability to differentiate into various cell lineages, stem cells are also being tested for efficacy in regenerative medicine. An additional characteristic of the totipotent stem cells is the ability to incorporate into a developing mouse embryo when injected into a blastocyst (Bradley et al. 1984). The stem cells participate in the development of the host embryo to produce chimericism in the resulting mouse. The production of mouse chimeras, an important step required to generate genetically modified mice, can alternatively be derived by aggregating ES cells together with a 2.5-day-old morula-stage embryo as well (Nagy and Rossant 2000; Eakin and Hadjantonakis 2006). If truly undifferentiated, the cells derived from an ES cell clone will be established in the germline of a chimeric mouse. Characteristics of the ES cell, such as coat color, can then be transmitted from the germline into the progeny of the chimeric mice. Thus, an ES cell clone that has been mutated in tissue culture can then be used to eventually derive a mouse containing this genetic modification.
Important clues about ES cell culture were originally derived by observations of teratocarcinomas. Grafting of early embryos ectopically into a histocompatible host leads to experimentally derived teratocarcinomas from which embryonic carcinoma (EC) cell lines could be cultured (Solter 1970; Stevens 1970). EC cells can differentiate into all 3 types of precursor tissue necessary for the development of a mouse, namely the ectoderm, mesoderm, and endoderm. Maintaining the EC lines on feeder cells of transformed mouse fibroblasts, however, helped to prevent differentiation (Martin and Evans 1974; Martin and Evans 1975). EC cells were determined to share homology and appearance with preimplantation-stage embryos (Evans and Kaufman 1981). The knowledge gained by studying EC cells, including the use of feeders to prevent differentiation, allowed for the eventual isolation of stem cells from the mouse embryo. Initial isolation of ES cells was performed by either explanting whole blastocysts (Evans and Kaufman 1981) or by isolating the inner cell mass with immunosurgery (Martin 1981). In the two decades after the initial isolation of these stem cells, more the 1,200 scientific papers have been published with mouse ES cells as a research emphasis (Downing and Battey 2006)
One very important factor that aided in the culture of ES cells was myeloid leukemia inhibition factor (LIF). This cytokine was initially given the name DIA for differentiation inhibitory activity and was isolated from various sources as a soluble factor capable of inhibiting differentiation in both ES and EC cells. Eventually DIA was cloned and characterized as the molecule LIF (Williams et al. 1988). LIF is a multifunctional cytokine that acts as a hemopoietic regulator. In addition, LIF is expressed by the trophectoderm of a developing embryo. Embryonic stem cell and embryonic carcinoma cell lines were shown to display high-affinity receptors for LIF. When highly purified recombinant LIF was added to cultures, more than 95% of the resultant colonies displayed a stem cell phenotype of compact colonies. In contrast, cells cultured in normal culture medium eventually differentiated to form colonies of large flat differentiated cells. LIF-mediated cell signaling, particularly through the activation of STAT3, plays a key role in promoting ES cell propagation while blocking cellular differentiation (Burdon et al. 1999). Other refinements to the ES cell culture media included the addition of beta-mercaptoethanol and nonessential amino acids, which are useful to maintain the viability of the cultured stem cell (Robertson 1987).
Recently, key transcription factors have been identified that may be responsible for maintaining pluripotency in stem cells. The combination of 4 transcription factors, Oct-3/4, Sox2, c-Myc, and Klf4, was shown to be able to reprogram mouse embryonic fibroblasts (MEFs) into a stem cell-like state (Maherali et al. 2007; Okita et al. 2007; Wernig et al. 2007). Oct-4, in particular, has been seen as a key regulator of pluripotency, and high expression of this transcription factor is characteristic of undifferentiated stem cells (Niwa et al. 2000). Upon retroviral infection of these transcription factors, MEFs were transformed into induced pluripotent stem (iPS) cells that behave similar to ES cells. After proper selection, these iPS cells could populate a chimeric mouse and be transmitted through the germline. The expression of the transcription factor Nanog, a downstream target of Oct3/4 and Sox2, is also critical for the maintenance of pluripotency in stem cells. Nanog deficiency in mice results in the inability for the inner cell mass to form an epiblast (Mitsui et al. 2003). Therefore, expression of Nanog is also related to maintenance of pluripotency. Cell surface changes can additionally be studied to determine if the stem cell cultures are undergoing differentiation. For example, changes in the expression level of stage-specific embryonic antigens (SSEA) are useful in determining if stem cells are undifferentiated. Differentiation of murine ES cells results in the loss of SSEA-1 expression and an increase in both SSEA-3 and SSEA-4 (Solter et al. 1979). This shift, however, is exactly opposite to human ES cells. Lastly, a high level of alkaline phosphatase is also a good indicator that the stem cells are in an undifferentiated state (Pease et al. 1990).
Historically, the most common ES cell lines were derived from the 129sv inbred mouse strain. The 129 mouse was often used to develop stem cells because this strain was commonly used to study embryonic development and was the favored strain in the derivation of EC cells. ES cells obtained from the 129 strain were unusually adept in colonizing and competing with cells from the inner cell mass of blastocysts. Commonly used 129-derived ES cell lines include ES-D3 (Doetschman et al. 1985), J1 (Li et al. 1992), and R1 (Nagy et al. 1993). Initially only 129 strain mice were conducive or “permissive” for establishing ES cells in vitro. As cell culture conditions were improved to favor proliferation and reduce differentiation, stem cell lines could be maintained in culture for more passages and ES cells could be isolated from previously “nonpermissive” strains, such as from C57 mice (Ledermann and Bürki 1991; Köntgen and Stewart 1993) and BALB/c mice (Noben-Trauth et al. 1996; Baharvand et al. 2004).
With the discovery of homologous recombination, scientists quickly realized that ES cells would be useful for gene targeting experiments. A targeting construct is delivered into a stem cell where homology arms in the vector align with a targeted genetic locus to introduce a gene modification. While not exactly the most efficient technique for producing homologous recombination events, electroporation became the preferred and easiest way for mass delivery of targeting constructs into a large number of stem cells (Vasquez et al. 2001). Targeting constructs usually contain a positive drug selection maker to enrich for clones that have successfully taken up the vector. The neomycin phosphotransferase (neor) gene is a well-known positive selection marker that confers resistance to the drug neomycin sulfate (G418) in tissue culture. In addition, gene targeting vectors also generally include a negative drug selection marker outside of the homology arms to select against random integrants (Mansour et al. 1988). For example, the HSV thymidine kinase (HSV-tk) gene is a widely used negative selection marker that phosphorylates drugs like gancyclovir or FIAU (1-[2′-deoxy-2′-fluoro-β-D-arabinofuranosyl]-5-iodouracil) into cytotoxic compounds. Clones with random integration of the complete vector are thereby excluded upon drug treatment. Thus, after electroporation of the targeting construct, the stem cells are plated onto culture dishes and treated for about 10 days under drug selection to select for clones that have undergone a true homologous recombination event. Once a stem cell with the designated genetic alteration is identified, the ES cell clone is then usually expanded and injected into a blastocyst to derive a chimeric mouse containing the cells with targeted mutation. The mice produced from mating chimeric mice with the wild-type mice will be either wild-type or heterozygous for the targeted mutation, and these heterozygous mice must be bred to homozygosity.
Homozygous null ES cells can also be derived in tissue culture by treating a properly targeted clone to increased positive drug selection pressure. For example, clones with a targeted insertion of the neor gene can be treated with a high concentration of G418 to force the replacement of the wild-type gene for the mutated copy (Mortensen et al. 1992). The high concentration of G418 seems to force another homologous recombination event to accommodate for the increased drug selection pressure. Null stem cells can then be characterized in culture to determine if gene inactivation affects the differentiation into various cell lineages. The homozygous null ES cells can also be injected into a mouse blastocyst to make a chimeric mouse. In the chimeric mouse, the development of the null cells can then be followed to determine if the affects of gene inactivation are cell autonomous.
In addition to use in gene targeting, ES cells have been used as a vehicle for transgenesis. ES cells can be genetically modified in culture through transfection of a transgenic construct. This ES cell route to making a transgenic mouse provides researchers with a means to screen for the desired genetic alteration before producing an animal model (Gossler et al. 1986). Along with electroporation or calcium phosphate transfection of transgenic constructs, viruses have also been a commonly employed for gene transfer into ES cells. One of the first experiments in genetic manipulation of ES cells involved the delivery of exogenous DNA through a retroviral vector (Robertson et al. 1986). The modified ES cells were then used to create genetically altered mice. Transduction of ES cells has been performed with adenoviral and lentiviral vectors as well (Smith-Arica et al. 2003; Kosaka et al. 2004). Gene transfer through viral vectors has sometimes provided better transduction efficiency and less cytotoxicity than electroporation.
Transgenesis using gene trap vectors is an alternative means of making knockout mice, although via indiscriminant gene inactivation (Gossler et al. 1989; Stanford et al. 2001). Unlike gene targeting, gene trap vectors are not site directed through homology arms and are therefore only a random means to disrupt a gene. Gene trap vectors generally consist of a promoterless reporter gene like lacZ (β-galactosidase) whose expression is dependent on random integration into a functioning gene. A splice acceptor is also commonly placed preceding the lacZ gene to ensure expression when integrated within a gene intron. Additionally, a gene trap vector usually will contain a neor gene for positive drug selection.
When planning to make genetically altered mice, careful consideration should be made in choosing the mouse strain of ES cell to use for experimentation. Most stem cells are derived from the 129 strain of mice because, historically, these mice were commonly used for the derivation of EC cell lines. However, extensive genetic variation has been reported between the 129 mouse substrains and the ES cells derived from these mice (Simpson et al. 1997). Additionally, the 129 strain of mice tend to have reduced fertility as compared with other mouse strains. Lastly, some reports indicated that this background displays some abnormalities in anatomy, immunology, and behavior (Seong et al. 2004). Still, 129 cells are easier to propagate in vitro when compared with stem cells from other mouse strains. In addition, stem cells from other mouse strains do not populate a blastocyst as efficiently and are less capable of leading to germ line transmission. This is particularly seen with higher-passaged stem cells (Schoonjans et al. 2003). Therefore, of the thousands of targeted mice described in the literature, most were created using the 129 mouse strain background. For example, only 38 targeted mice have been developed with a C57BL/6 background even though 2 C57BL/6-derived ES cell lines have been available for more than a decade (Seong et al. 2004). To get a pure breeding background, though, requires at least another year of backcrosses. Therefore, trying a stem cell from strains other than 129 can sometimes be a worthwhile option. For example, stem cells from the BALB/c mouse were derived because this strain is commonly used for experiments in immunology (Dinkel et al. 1999). The C57 strain of mice is also favored for studying various phenotypes since this strain has been well characterized. Therefore, C57BL/6 stem cell lines have been developed, and more genomic DNA libraries have recently been constructed from C57 strain mice to help enable investigators to develop knockout mice with this background. With this strategy, C57-derived ES cells (e.g., E14 or Bruce4) are microinjected into C57 donor blastocysts and foster mothers. Coat color cannot be used as a marker for chimerism since the pups would have a black coat color regardless of recombined ES cells or native ES cells. Recently, C57 ES cells have been developed with a tyrosinase mutation that imparts a white color (albino), allowing coat color assessment of chimerism. Although the C57-derived ES cells require more fastidious care than the 129-derived cell lines, the targeting frequency between these two strains is roughly similar (Seong et al. 2004). In addition, while the 129-derived ES cells are better able to proliferate in a host blastocyst, the capacity to transmit the targeted event through the germ line is not very different between the two strains. Regardless of which strain of ES cell lines is used, care must be taken to ensure that these ES cells remain totipotent until they are transferred to a host blastocyst and foster mouse.
BASIC PROTOCOL 1
Culturing of primary mouse embryonic fibroblasts and derivation of mouse embryonic stem cell feeders
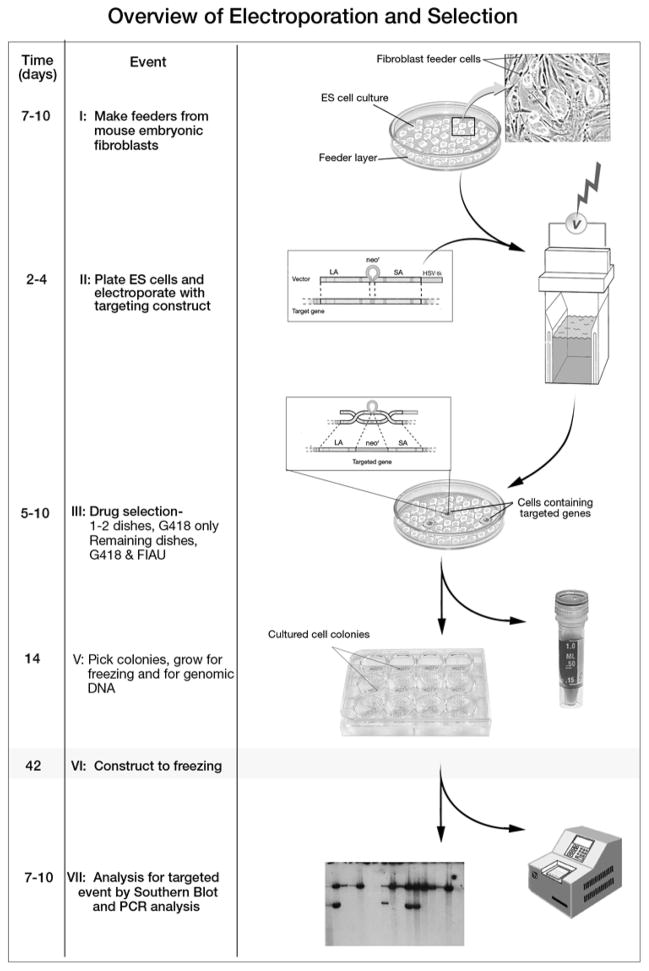
For a gene targeting experiment, mouse embryonic stem cells in culture need to be maintained in a pluripotent stage until they are injected into mouse blastocysts. Usually, to maintain these conditions, a feeder cell layer is used to provide an environment that helps to prevent differentiation. The feeder cells secrete unknown growth factors that help preserve pluirpotency. Feeder cells are produced from either STO embryonic fibroblasts or primary mouse embryonic fibroblasts (PMEFs). PMEFs are typically employed in gene targeting experiments and are derived from E14.5 day old embryos. The embryos are minced, and the dissociated cells are placed into culture. The adherent fibroblasts are then usually passaged a few times to obtain an adequate number of cells to be frozen in aliquots. PMEFs can also be purchased from commercial sources, which can save time and cage space. One important point is that the PMEFs should be resistant to whichever positive selection cassette used within the gene targeting construct (e.g., neomycin). Otherwise, this feeder layer will not support ES cell growth once the drug selection is introduced. PMEFs can be generated from the embryos of an established knockout mouse that bears the same drug resistance cassette. Fibroblasts derived from the knockout mice should have the necessary resistance cassette integrated into their genome. Feeder cells need to be growth inhibited before co-culture with ES cells. Typically, the cells of mitotically inactivated by either treatment with the drug mitomycin C or through γ-irradiation. If using mitomycin C treatment, be sure to wash the cells thoroughly since this drug can cause differentiation of the stem cells. To enhance the growth conditions needed to maintain pluripotency, the culturing of ES cells on PMEFs is usually supplemented with the use of gelatin coated plates and the addition of LIF (leukemia inhibitory factor) to the stem cell medium. LIF is a cytokine that helps to prevent differentiation of the cells. While ES cells can be maintained with LIF in the absence of PMEFs, an increased tendency towards differentiation will develop with long term culture of these cells (Nichols et al. 1990). This especially seems to be the case with ES cells that have undergone many passages. A brief illustration depicting the critical steps in the generation of targeted embryonic stem cells is shown in Figure 1.
Figure 1.
Overview of Electroporation and Selection. The left column shows the approximate number of days at each critical step in the preparation of targeted embryonic stem cells. The middle column describes key procedural events. The last column is an illustration of each critical event. Note that in stages II through V embryonic stem cells should remain as close to pluripotent as possible. Once the cultured ES cells have been plated to the tissue culture plate depicted in step V, they may become differentiated. These cells will used for DNA screening as depicted in step VII. In step VII, the left panel represents an autoradiographic film image of a Southern blot. Southern blot is the definitive test for detection of a targeted event. Many investigators first screen all clone DNA samples by PCR followed by confirmation with Southern blot. A strategy for differentiating between the knockout allele and the wild-type allele should be worked out in advance of initiating a targeting experiment.
Materials Required
Equipment
Laminar flow hood- Class II/Type A/B3 Model #NU-407-400 (NuAire)
Tissue culture incubator, water-jacketed (Thermo-Forma)
Dedicated centrifuge able to achieve 4 °C (Beckman-Coulter Allegra X-22R model)
Inverted microscope with phase-contrast filter, model ID03 (Carl Zeiss)
Curved iris scissors (Roboz surgical instruments)
Single-edged razor blades
Sterilizer pouches to autoclave instruments (Fine science tools)
Hemocytometer or Cell Counter
37°C Water Bath
Pipette Device (Eppendorf)
Pipettors
Reagents
10-cm2 tissue culture-grade dishes (Fisher Scientific)
6-cm2 tissue culture-grade dishes (Fisher Scientific)
Sterile Erlenmeyer Flasks (Corning)
Serological Pipettes (1 ml, 2 ml, 5 ml, 10 ml, 25 ml, 50 ml)
Pipette Tips
Sterile 15 ml and 50 ml centrifuge tubes
70% Ethanol
Trypsin-EDTA, 0.05% (Invitrogen)
Mitomycin C powder, 2-mg vial (Sigma-Aldrich)
Dulbecco’s Phosphate-Buffered Saline without Ca2+ or Mg+ (DPBS) (Cellgro)
1-liter filter unit, 0.22-μm pore size (Millipore)Embryonic Feeder Media (EFM)
500-ml bottle DMEM (Dulbecco’s Modified Eagle’s Media with high glucose and phenol red) (Invitrogen)
50 U/ml penicillin/streptomycin solution 1000X (Invitrogen)
6 mmol/L L-glutamine solution, 100X (Invitrogen)
10% fetal bovine serum (FBS, Gemini Bio-Products)
2X cell freezing media
2 ml DMSO (Dimethyl Sulfoxide, Sigma Chemical
2 ml Fetal Bovine Serum (FBS, Gemini Bio-products)
6 ml EFM
Filter sterilize FBS and EFM before adding DMSO
Gelatin, porcine skin, type A, 0.2% in tissue culture-grade water, sterile (Sigma-Aldrich)
Dissolve 2 g of gelatin in 2000 ml of water then sterilize by using an autoclave.
Procedure: Derivation of Primary Embryonic Fibroblasts (PMEFs)
Mate male mice harboring a positive resistance cassette to a similar female mouse or heterozygous female.
Observe the copulation plug and wait until the embryos are day 13.5 to 14.5
Prepare three tissue culture dishes filled with sterile DPBS along with sterile, autoclaved scissors and forceps.
Euthanize the mouse by CO2 asphyxiation, cervical dislocation, or which ever method has been approved by your institute Animal care and use committee.
Wipe the fur surrounding the abdominal cavity with 70% alcohol to clean and prevent fur and dander from entering into the abdominal cavity.
Make a vertical incision down the abdominal cavity and locate the uterus. The uterus should be swollen with embryos, resembling a sac with many bulges.
Detach this sac from both ends of the abdominal cavity with fine tipped scissors and place into one dish filled with 15 ml sterile DPBS. Move the uterus around in the dish to wash off some of the blood.
Cut open the sac releasing the embyos into the DPBS.
Transfer the embryos to a new clean dish of DPBS.
Hold each embryo with forceps, scoop out the liver, and decapitate the embryos with the single-edged razor blades.
Rinse the carcasses and transfer to a new dish of DPBS. Try to get rid of as many red blood cells as possible.
Prepare one tissue culture dish with 15 ml trypsin-EDTA solution. Place this dish inside a clean laminar flow hood. Place the decapitated embryos into the trypsin dish and quickly mince the tissue with the forceps and curved scissors until the tissue can be sucked up into a 10 ml serological pipet. Pipet minced tissue up and down into the same dish 6–7 times.
Add another 5 ml trypsin solution, and continue to disperse the tissue with a 5 ml pipet.
Place this dish in the incubator for 10 minutes.
Transfer the contents to a sterile 50 ml conical tube to allow the undigested tissue to settle at the bottom of the tube. This should take about 2 minutes.
Decant the supernatant to a new, empty tube and bring the volume up to 50 ml with EFM.
Centrifuge this tube at 1000 rpm for 5 minutes at 10°C. Then, discard the supernatant.
Resuspend the cell pellet in 50 ml fresh EFM. Transfer 10 ml per dish to a sterile tissue culture dish.
Allow the fibroblasts to attach overnight in the incubator. The next day replace the media from each dish with EFM. At this point the mouse embryonic fibroblasts (PMEF) will be passage 0.
Often passage 0 fibroblasts are “split” into several more dishes to permit preparation of a bigger batch of frozen PMEF.
Procedure: Expansion of PMEFs for frozen stock
To split the PMEF, aspirate the EFM from each plate. Add about 5 ml DPBS to each dish then aspirate the DPBS off of the dish. This wash reduces any dead cells and removes serum from the media, which can inhibit the action of trypsin.
Add 2 ml trypsin to the dish and place in the tissue culture incubator at 37°C for 3 minutes.
Gently tap the side of the tissue culture dish and check that the PMEF have detached from the bottom of the dish.
Add 3 ml EFM to the dish to stop the action of trypsin. Pool all the dishes that were trypsinized and bring the volume up to the amount necessary for 10 ml cells per dish. For example a splitting ratio of 1:3, would require 10 ml cell suspension to 20 ml EFM.
The passage number of the newly split cells would now be passage 1 (P1). Depending upon the density of the PMEF, the dishes will be confluent in about 4 or 5 days.
We generally freeze down aliquots of MEF at passage 3. Cell suspensions should be made as described in steps 22 to 24. Count a small aliquot of the cell suspension with a hemocytometer or automated counting device. For a hemocytometer 10 μl of cell suspension is added to the chamber and will flow by capillary action to the counting grid. Count the number of MEF in each outer square. Divide this total number by 4 for the average number per square. Multiply this value by 1 × 104 (the cubic volume) to get the number of cells per ml. The number of dishes prepared for freezing should be sufficient for 2 × 107 cells per 0.5 ml cells. Combine 0.5 ml of the cell solution with 0.5 ml 2X freezing media per freezer vial. The final concentration will be 1 × 107 cells per vial.
Representative vials should be tested for mycoplasma and other potential mouse pathogens before a stem cell experiment is initiated.
PMEF should not be passaged more than about passage 6, otherwise the capacity of the feeders to provide the critical cytokines might be compromised.
Procedure: Expansion of PMEFs for production of feeders
Pre-warm 9 ml of EFM (after filter-sterilizing the media) to 37 °C.
Thaw a vial of PMEF in the 37 °C water bath until the cells are half thawed. Clean the exterior surface of the freezer vial with 70% ethanol. Take one-half of the freezer vial’s volume (generally a total of 1 ml) and transfer to a centrifuge tube with the pre-warmed EFM. Then transfer a portion of the pre-warmed medium to thaw the remaining PMEF stock. This ensures the rapid thawing of the PMEF and the quick dilution of the freezing buffer containing DMSO. Aspirate the entire contents of the freezer vial back into the centrifuge tube.
Centrifuge the cells at 1800 rpm (400g?) for 5 min at 4 °C and aspirate the supernatant.
Resuspend the cell pellet in 20 ml EFM and then dispense the media and cells at 10 ml per dish to 2 × 10-cm2 dishes.
Transfer the dishes to a tissue culture incubator at 37 °C with 5% CO2.
After 3 to 4 days depending upon the density of cells, the PMEFs should be confluent.
Once confluent, aspirate the media from each dish and wash the cells with DPBS to cover the cells. Aspirate the DPBS, and then cover the cells with cold trypsin (2 ml per dish).
Incubate for 3 min at 37 °C until the PMEFs to detach from the dish.
Neutralize the trypsin by adding EFM (3 ml per dish). Pipette up and down several times to detach the cells and make a single cell suspension.
Pool the media from both dishes into a centrifuge tube. Wash off any remaining cells on the culture dishes with an additional 5 ml EFM and add to the pooled media.
Centrifuge at 1800 rpm for 5 min at 4 °C.
Resuspend the cells and dilute in a sterile Erlenmeyer flask with EFM to a solution of 150 to 200 ml total. Add 10 ml of the cell solution to the 10-cm2 dishes to make 15–20 plates of PMEFs.
Procedure: Treatment of PMEFs with mitomycin C (MMC)
Treat about 40 × 6 cm2 dishes with 4 ml of gelatin for about 30 min at room temperature in a sterile laminar flow hood. Then discard the gelatin, and allow the dishes to dry to provide a surface for the PMEF to attach.
Replace the EFM media in 15 dishes (~5 dishes at a time) of PMEF with EFM containing MMC at 0.01 mg/ml. [Dissolve 1 vial of MMC with 2 ml of DPBS and then transfer all of the contents into 198 ml of EFM – 2mg MMC/200 ml EFM] Note: The MMC is a potent toxin, so care must be taken to handle it carefully and to dispose of it as chemical waste. Reserve the remaining 5 dishes of PMEF for MMC treatment later in this protocol.
Place the MMC-treated PMEF in the incubator for 3 hours to permit metabolic arrest of the PMEF.
Aspirate the EFM+MMC into the appropriate chemical waste container. Wash each dish with 3 washes of 5 ml DPBS to ensure total removal of the MMC. Treat the washes as hazardous chemical waste.
Add trypsin (2 ml per dish) to cover the fibroblasts in each dish and incubate for 3 min at 37 °C.
Neutralize the action of trypsin with 3 ml EFM, and pool the volume (5 ml) from each dish into a 50-ml centrifuge tube. Wash each stack of 5 dishes with an additional 5 ml of EFM and add to the pooled cell suspension.
Centrifuge at 1800 rpm for 5 min at 4 °C.
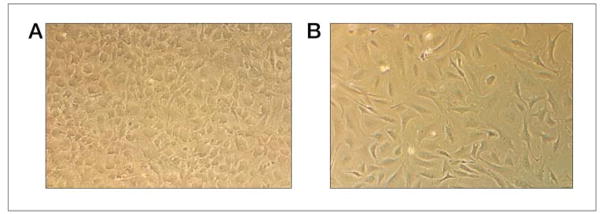
Aspirate the media, and re-suspend the pooled cell pellets in a convenient concentrated volume (such as 1/10th the original volume, ~15 ml). Count a small aliquot of the cell suspension using either a hemocytometer or an automated cell counting device. Dilute the cells in EFM to a concentration between 0.8–1.0 × 105 cells per ml. This concentration is ideal to form a monolayer for the ES cells to grow. Transfer this cell suspension to at least 25 gelatin-treated 6-cm2 dishes. Within the next day or two, the feeders will resemble a cobblestone-like appearance (see Figure 2). These feeders will secrete factors in the media, which will provide an environment along with LIF (leukemia inhibitory factor) for the mouse ES cells to grow.
Figure 2.
Mitotically inactivated ES cell feeders and PMEF. Panel A is primary mouse embryonic fibroblasts that have been treated with Mitomycin C. The treatment arrests the cell division and produces cytokines, which inhibit differentiation of ES cells. Note that the cell bodies appear swollen and resemble a cobblestone-like appearance. Panel B shows PMEF that have not been treated with Mitomycin C. These fibroblasts have a elongated and sickle-shaped appearance.
BASIC PROTOCOL 2
Culturing mouse embryonic stem cells
Materials Required (in addition to the items listed in Basic Protocol 1)
Reagents
Mouse embryonic feeders from Basic Protocol 1
Pluripotent mouse embryonic stem cells (lower passage preferred – usually no more than 20 passages)
Examples Include:
R1 (available from American Type Culture Collection, ATCC) [129 strain-based]
W4129/S6 (Taconic) [129 strain-based]
Bruce4 [C57BL/6 strain-based]
Pluristem B6 albino (Millipore) [C57BL/6 strain-based]
Embryonic stem cell media (ESM)
500-ml bottle of DMEM (Invitrogen)
15% fetal bovine serum (FBS), embryonic stem cell qualified (Gemini Bio-Products)
50 U/ml penicillin/streptomycin solution, 1000X (Invitrogen)
6 mmol/L L-glutamine solution, 100X (Invitrogen)
90 μM/L 2-β-mercaptoethanol, tissue culture grade (Invitrogen)
100 μM/L nonessential amino acid solution (Invitrogen)
1000 U/ml leukemia inhibitory factor (LIF), (Chemicon-Millipore)
(Note: ESM should be used in less than 4 weeks before glutamine begins to degrade. Lots of FBS should be screened to ensure that the serum is able to maintain stem cell cultures in an undifferentiated state. Since FBS can contain growth factors that may encourage differentiation, a serum-free formulation, KnockoutSR, is available from Invitrogen,)
Procedure: Plating embryonic stem cells
Prewarm 9 ml of ESM (after filter-sterilizing the media) to 37 °C. Thaw 1 vial of R1 ES cells (1 ml) in a manner similar to step 1 for PMEF in order to dilute out the DMSO cryoprotectant. Generally, the vial should contain about 1 × 106 cells. If the vial contains too few cells then the growth maybe inhibited. If the vial contains too many cells then differentiation may occur as the cells will be too close to each other.
Centrifuge the ES cells at 1800 rpm for 5 min.
Gently resuspend the cells in 12–16 ml ESM.
Aspirate the EFM from 3–4 feeder dishes created in Basic protocol 1, and replace with 4 ml ES cell suspension per dish.
Once ES cells have been plated onto feeders, they must be re-fed daily with LIF-containing ESM to help maintain pluripotency. Depending upon the freezing density of the ES cells, the plate should be ready for harvesting in 2–3 days.
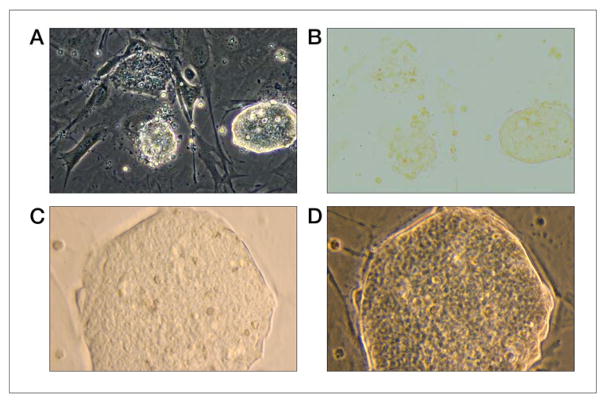
Under suitable conditions, the ES cell colonies should have very smooth, sharp borders with respect to the underlying feeders and have a dome-like, almost shiny appearance (see Figure 3 panel A and D). Stem cells maintained in culture need to be split every two to four days to prevent differentiation. Differentiation can occur if the stem cells are plated at either too low or too high of a density. The stem cell colonies should not be allowed to become large and overgrown and the ES cells within these cultured colonies should appear indistinguishable. Differentiation is generally seen when stem cell colonies become large and flattened with jagged borders.
Figure 3.
Panels A and D are images of embryonic stem cells taken with phase contrast filter. Panels B and C are bright light images. Panel A shows embryonic stem cells under drug selection pressure. The far right colony appears to be surviving drug selection (note the sharp, bright border. The other two colonies (rough border) probably would not survive drug selection. Panel D shows a high magnification of a good embryonic stem cell colony. The border appears very sharp and the mass of embryonic stem cells within it appears indistinguishable from each other.
BASIC PROTOCOL 3
Introduction of plasmid DNA into pluripotent mouse stem cells
Materials Required (in addition to the items listed in Basic Protocol 1 and 2)
Equipment
Gene Pulser Xcell Electroporation system with capacitance extender (Biorad) Electroporation cuvette 0.4-mm gap (Biorad)
Reagents
0.25% Trypsin-EDTA (Invitrogen)
Electroporation buffer (EB)
10% Hank’s balanced salt solution (HBSS), (Invitrogen)
20 mmol/L HEPES buffer solution (Invitrogen)
1 mmol/L sodium hydroxide (NaOH) in water, (Sigma-Aldrich)
110 μmol/L 2-β-mercaptoethanol, tissue culture grade (Invitrogen)
Procedure: Preparation of targeting construct and embryonic stem cells for electroporation
Refeed the ES cell culture dishes with ESM to encourage cell growth before electroporation.
The targeting vector should be linearized with the appropriate restriction enzyme then purified and dried, generally through ethanol precipitation. The plasmid should be free of any contaminants such as ethanol and phenol.
Dissolve 25 μg of targeting vector in 25 μl of EB to make a final concentration of 1 μg/μl.
Aspirate the media from the ES cell dishes, and wash with 2 ml of DPBS per dish. Aspirate the DPBS and then cover the cells with 1 ml of 0.25% trypsin per dish. Incubate for 3 min at 37 °C.
Inactivate the trypsin with 4 ml of ESM per dish. Pipette the cells vigorously to dissociate the stem cell colonies into a single cell suspension. Pool the cell media mixture from all the dishes and collect into a centrifuge tube. Wash any residual cells remaining on the culture dishes with an additional 5 ml of ESM and add to the pooled cell suspension.
Centrifuge the ES cells at 1800 rpm, at 4 °C for 5 min.
Aspirate the supernatant and resuspend in 5–10 ml ESM. Count a small aliquot of the ES cells using either a hemocytometer or an automated cell counting device (If using the hemocytometer, the ES cells will appear rounder and smaller than the arrested feeder cells) Select a volume of cell suspension that will ensure 1 × 107 cells. Some investigators use Trypan blue to differentiate between dead and live cells. The dead (blue) cells will not be able to remove the trypan blue and should be excluded from the count if using a hemocytometer.
Centrifuge the ES cells at 1800 rpm, at 4 °C for 5 min. Aspirate the supernatant and resuspend the cell pellet in 0.8 ml of EB. Some investigators have used sterile Phosphate buffered saline on ice. Transfer the cell solution to a sterile 1.5-ml tube.
Add the targeting construct to the tube containing the ES cells, and gently mix the DNA with the ES cells. At this stage, it is important to have a single cell suspension. Transfer the mixture to the 0.4 mm electorporation cuvette (try to avoid forming bubbles).
Place the cuvette into the shock pod of the electroporator. The metal plates on the cuvette should be in contact with the metal plates of the shock pod. Electroporate the cell suspension at the appropriate setting. Usually most cells like R1 use a setting of 230 V, 500 μf, and an electrode gap of 0.4 cm with an exponential decay. With these settings, the time constant should be between 5.6 to 8.0 ms, depending upon the solutes in the media and the cell concentration. After the shock, bubbles or foam at the top of the cuvette will be apparent, signaling that current has passed through the solution. Optimal settings will vary depending upon ES cell type and must be determined experimentally.
After electroporation of the cells, leave the cuvette containing the electroporated cells at room temperature for 10 min. For mammalian cells, it is not critical to chill the cells before or after electroporation.
Transfer the entire contents of the electroporation cuvette to 200 ml of ESM in a disposable, sterile, Erlenmeyer flask, resulting in a concentration of 5 × 104 cells per ml plated onto feeder dishes. Wash the cuvette with about 0.8 ml of the ESM to retain as much of the electroporated cells as possible. As a result of this shock, many of the cells will die off and will be seen floating in the media. With the initial cell density of 1 × 107 cells, enough cells should survive to undergo drug selection.
BASIC PROTOCOL 3
Selection of drug-resistant cells, freezing drug-resistant cells, and preparation of cell lysates for analysis
Materials Required (in addition to the items listed in Basic Protocol 1 and 2)
Equipment
Flame sterilizer (Fireboy Plus, Integra Biosciences)
Forceps
Mr. Frosty slow cool chamber (Nalgene) with isopropanol (Fisher Scientific)
−80 °C freezer (Thermo-Forma)
Liquid nitrogen freezer (Thermo-Forma)
Pipettor from 20 μl to 200 μl to mix cell suspensions + sterile tips (Rainin)
Pipettor from 200 μl to 1000 μl to mix cell suspensions + sterile tips (Rainin)
Repeat pipetting device from 100 μl to 1000 μl (useful but optional, Eppendorf)
Optional- Light Touch System (Rainin) requires less hand pressure and reduces fatigue
Reagents
0.25% Trypsin-EDTA (Invitrogen)
24-well tissue culture plates (Fisher Scientific)
12-well plates (Fisher Scientific)
2-ml liquid nitrogen cryovials (Nalgene/NUNC)
Positive selection agent: G418 sulfate (Invitrogen) analog of neomycin
Add 1 gram of G418 sulfate powder (743 μg/mg activity) to 10 ml tissue culture-grade water to make a stock solution (refrigerate at 4 °C. Supplement a bottle of ESM (~600 ml) with 2.1 ml of the G418 solution. The final concentration will be 350 μg/ml. Filter-sterilize ESM+G418 before use. The ideal G418 concentration must be determined experimentally for each ES cell strain by performing a “kill” curve. Briefly, naïve ES cells are plated and fed ESM containing increasing amounts of G418 (usually 50 to 100 μg per ml increments). Select the lowest concentration that “kills” all of the naïve ES cells within 3–4 days of selection pressure.
Negative selection agent: FIAU (1-[2′-deoxy-2′-fluoro-β-D-arabinofuranosyl]-5-iodouracil) (Moravek Biochemicals)
The FIAU comes in 1mg vials. It is convenient to first dissolve the entire contents of the vial in 1 ml 50% ethanol/50% water. Add 1 ml of the water/ethanol mix and the FIAU will dissolve slowly. Ensure that no precipitate is present at the bottom of the vial. The resulting solution will be 270 μmol/L. Transfer the entire contents to 9 ml of ethanol/water mix. Aliquot to several tubes and add 444 μl of this stock to 600 ml of ESM for a final concentration of 0.2 μmol/L. Filter-sterilize this along with the G418 solution to make ESM+G418+FIAU.
2 X ES cell freezing media
90 ml of ESM
30 ml of fetal bovine serum, FBS, ES cell qualified (Gemini Bio-Products)
Filter sterilize, 0.22-μm pore size (Millipore)
Add 30 ml of dimethyl sulfoxide, DMSO, (Sigma-Aldrich) Addition of the DMSO to the filter unit sometimes clogs the membrane.
Cloning cylinders 8 mm X 8 mm (Bellco Glass)
High-Vacuum Grease (Fisher Scientific)
Dish of glass cloning cylinders (Prepare cloning cylinders with grease before picking stem cells)
Lightly and evenly grease the bottom of a glass dish
Fill the dish by placing the flat bottom of the cloning cylinders into the grease
Cover and autoclave
Procedure: Drug selection and freezing clonal cells
Drug selection for resistant clones should begin within 20 hours of electroporation of targeting vector DNA into ES cells, which helps to reduce the population of nontargeted ES cells.
The majority of dishes should be treated with ESM supplemented with G418 and FIAU (i.e., positive and negative drug selection). One or two dishes should be treated with ESM and G418 only (i.e., only positive drug selection) to check for the degree of random targeting vector insertion (targeting vector inserted randomly in the genome should include the HSV-TK cassette and would be toxic to the ES cell colony).
Replace the medium with ESM+G418+FIAU for 4–5 days of daily treatment followed by ESM+G418 only for 3–4 days.
After this period, pinpoints of circular, white, drug-resistant colonies should be visible without a microscope. Lift each dish and circle the under side of the dish containing candidate colonies for expansion. Colonies that are starting to differentiate will sometimes appear more flat with a diffuse “haze” around them. These colonies should not be picked whenever possible.
The day before picking colonies, prepare 24-well feeder plates to receive the targeted clones for expansion. Prepare the feeders as described in Basic Protocol 1. The MMC treated feeders should be at a concentration of 0.8 to 1.0 × 105 cells per ml. Dispense 0.5 ml of the feeders per well on gelatinized 24-well plates. There should be enough plates and wells to receive at least 200 colonies.
The next day, replace the 0.5-ml volume per well in the 24-well plates with 1 ml of ESM before starting to pick the stem cell colonies from the 6 cm2 dishes.
Aspirate the medium from one 6-cm2 dish and replace with 1–2 ml of DPBS.
Aspirate the DPBS from the dish. Sterilize a pair of forceps with a flame and use the forceps to vertically lift the cylinders swiftly off the glass dish. The grease should be evenly distributed along the bottom of the cylinder. Place a greased cloning cylinder around a suitable colony and gently press onto the culture dish. Ensure that the cylinder is positioned around only a single colony so that no adjacent colonies are harvested in the process. When possible, try and avoid two colonies in close proximity to each other. In this case, one colony may “pass-on” neomycin resistance to the nearby colony, as both colonies would fit under the same cloning cylinder and would be picked together. This is the “bystander” effect.
Repeat these steps (flaming the forceps each time) until all good colonies have a cloning cylinder above them.
Dispense 200 μl of 0.25% trypsin in each cloning cylinder (use the repeat pipettor to dispense the trypsin over the cylinder). The vacuum grease will hold the liquid inside the cylinder, acting as a miniwell. Usually it is best to limit the number of dishes to 1 or 2 at a time to prevent over-trypsinizing the colonies.
Incubate the dish in the tissue culture incubator for 3 min at 37 °C.
After 3 min., use a 200 μl pipette and a sterile tip to pipette the trypsin up and down in the cloning cylinder. Pipetting is needed to disperse the colony clumps. If dissociated correctly, a single cell suspension should have been achieved through trypsinization.
Take the entire contents of each mini-well on the dish and dispense into one well of the 24-well plate feeders containing 1 ml of ESM. Repeat with new pipet tips each time until all the colonies have been transferred to the 24-well plates of feeders.
Replace the media in the 24-well feeder plates every day with 1.0 ml of ESM until the ES cells have grown sufficiently. Watch the stem cells daily. Be prepared to freeze the cells whenever the growing colonies in the well appear to be getting confluent or close to touching. It is probably a convenient point to prepare the 2X freezing media at this stage. Dispense 0.5 ml of 2X ES cell freezing media into each freezer vial and store at 4 °C. While an ample number of cells need to be grown for storage and freezing, ensure that the stem cell culture does not become overgrown and start to differentiate. The ES cell colonies should have very smooth, sharp borders (see Figure 3) and should not appear flat. If the media indicator appears orange to yellow, then the colonies are probably over confluent and may be ready to differentiate. These wells should be avoided whenever possible.
Prepare gelatinized 12-well plates with 1 ml of 0.2% gelatin per well. These plates will be needed for DNA testing of the potentially targeted clones.
The stem cells should be ready for harvesting within a few days after the colonies were isolated with the cloning cylinders. Examine each well daily to determine which colonies need to be frozen down. Mark the appropriately confluent wells on the 24-well plates to prepare for harvesting. The growth rate of the stem cell colonies will vary amongst wells on the tissue culture plate. Often, these differences can be due to the amount of cells recovered when picking the colonies.
Aspirate the media from the marked wells and replace with 1 ml of DPBS per well.
Replace the DPBS with 200 μl of 0.25% trypsin and transfer to the incubator for 3 min at 37 °C.
Neutralize the trypsin with 800 μl of ESM. The total volume in each well of the marked 24-well plates should be 1 ml.
Using a 1-ml pipettor with a sterile tip, pipette up and down to make a single cell suspension. It is often convenient to use pipettor that requires less force to pipette up and down, particularly when many clones are to be frozen down. We have used the LTS pipettors and pipet tips (Rainin) for this purpose.
Remove 500 μl of the cell suspension, and transfer it to the pre-filled freezer vial to make the volume 1 ml. Place the freezer vials in a slow cool chamber.
Transfer the remainder of the cell suspension (about 500 μl) to the pre-filled (with 500 μl ESM) well of the gelatinized 12-well plate. Transfer the 12-well plates to the incubator.
After collecting the stem cell freezer vials, transfer the vials to the slow-cool chamber into a −80 °C freezer (the temperature in the chamber should drop about 1 °C/min). Once the freezer vials are frozen at −80 °C, transfer the vials to a liquid nitrogen freezer for longer-term storage. Generally, these tubes should be stored in the vapor phase of the freezer, which minimizes the chance cracking of the plastic resulting in liquid nitrogen seeping into the freezer vials and cross-contaminating the samples.
Over several days, the ES cells in the 12-well plates should differentiate and overgrow, resulting in the color of the indicator in the media turning yellow. It is not necessary to maintain pluripotentcy in the 12-well plates, since the cells will only be used to purify genomic DNA. Genomic DNA from the yellowing clones can be purified (Laird et al. 1991) and used to determine whether a targeted event has taken place either by Southern blot or by PCR.
Additional comments
Alternate method
In general, one can expect a targeting efficiency (number of clones possessing the recombined allele divided by the total number of clones) of about 1 to 3 percent (Te Riele et al. 1992). In many cases, it is advantageous to have more than one targeted clone to increase the chances of obtaining good chimeric mice. The protocol described here can be very laborious, particularly at the steps where clones are picked and individually frozen. It is not uncommon to freeze 200 to 300 clones per project. Other investigators have developed a less laborious method that uses 96-well tissue culture plates and a multichannel pipette to reduce fatigue (Ramirez-Solis et al. 1993). In this method, feeders are plated onto 96-well tissue culture plates instead of 24-well plates. After drug selection, ES cell clones are scraped around the colony and plucked from the dish using a pipette tip and are then trypsinzed and transferred to corresponding 96-well feeder plates. When the cells are confluent and ready to freeze, a multichannel pipette is used to wash, add trypsin and media, and resuspend the cells. This method can generate many more clones and reduce the time involved and the resulting fatigue. Many investigators have used this method particularly if they use only positive selection when there is the concern over TK-induced sterility of transgenic males (e.g., conditional knockout mice where the exon has flanking loxP sites and an incorporated HSV-TK cassette) (Salomon et al. 1995) or when it is not feasible to include a negative selection cassette due to a lack of appropriate restriction enzyme sites. There are, of course, disadvantages using this “high-throughput” method. The chief disadvantage is that there is less cell lysate from the 96-well as compared to the 24-well method. Usually, there is only enough lysate to permit one Southern blot. Secondly, an entire plate must be harvested when the cells are ready to freeze if a multichannel pipettor is used, and the number of cells per well will vary.
Feeder-free system
The methods detailed above use mouse embryonic fibroblast cells that have been arrested; these feeders provide not only the cytokines necessary to maintain undifferentiated embryonic stem cells, but also a support for the stem cell colony to grow. In some cases where the population of dispersed embryonic stem cells is sparse relative to the number of feeder cells, it is difficult to differentiate between the wild-type genomic DNA from the feeder cells and the genomic DNA from the potentially null stem cells. In addition, it may be necessary to culture the PMEF for 1 or 2 weeks, adding time to any project. These problems led to identifying the particular growth factors in feeder cells that sustained self-renewal in embryonic stem cells. This possibility was especially useful in the propagation of primate stem cells where generation of feeders would be impractical. For a feeder-free system, LIF and the bone morphogenetic proteins (BMP-4) seem to be the only growth factors necessary for mouse ES cells to remain undifferentiated (Ying et al. 2003) without the presence of feeder cells. Millipore has developed a feeder-free kit for murine embryonic stem cell culture, and information for this kit can be obtained from their website (www.millipore.com).
Embryonic stem cell passage number
Occasionally, after targeted clones have been injected, chimeric mice are not produced, or chimeric mice are bred but the targeted ES cells do not contribute to the germline. One reason why appears to be the amount of time and passages spent in vitro. The steps necessary to generate and test new stem cell lines can be very long and require considerable skill. As a result, many facilities purchase stocks of these cells then freeze down large batches. If the passage number becomes very high, then the ability of these clones to contribute to the germline diminishes. One such study found that previously euploid (40 chromosome count in mice) ES cell clones, when passaged in vitro for more than 20 passages, rapidly became aneuploid (higher chromosome counts), and the capacity to contribute to the germline dropped to zero (Longo et al. 1997). Some facilities routinely screen new stem cell line batches by karyotyping chromosomal spreads, and some facilities offer this screening as a service. Karyotyping or g-banding naïve ES cell batches may help when troubleshooting problems with germline transmission.
History
The earliest attempts to culture mouse embryos were basically unsuccessful. In general, when a mouse blastocyst was placed in culture, the trophectoderm layer would grow into giant cells while the inner cell mass (ICM) would fail to proliferate (Cole and Paul 1965). Long-term cultures were eventually developed from mouse blastocysts, but over time, these would primarily consist of either epithelioid or fibroblastic cells (Sherman 1975).
Because of the initial problems in culturing blastocysts, many researchers turned to embryonal carcinoma (EC) cells to study embryonic development. Researchers discovered that when 1- to 7.5-day-old mouse embryos were transplanted into extrauterine sites in a host mouse, a teratocarcinoma would eventually develop (Solter 1970; Stevens 1970). EC cell cultures were then derived from these teratocarcinomas. Most of the EC cell lines were obtained through experiments with 129 mice, a mouse strain that was known to develop spontaneous testicular teratomas (Stevens and Little 1954). These cultured EC cells behaved like cells seen in an early embryo. In addition, the pluripotent EC cells could differentiate into all 3 primary germ layers and could be directed to form embryoid bodies in culture (Martin and Evans 1975). Unlike normal embryonic cells, however, the EC cells had an abnormal karyotype.
Studies into embryonic development were further augmented by the ability to generate chimeric mice. The cell-cell interactions that occur in a developing embryo could be studied by the injection of foreign cells into a host mouse blastocyst. R.L. Gardner (1968) was able to first show how the injection of cells into a blastocyst could produce mouse chimeras. This technique was then applied to show that EC cells could differentiate into various cell lineages to populate the developing chimeric mouse embryo (Brinster 1974; Papaioannou et al. 1975). The EC cells, however, were not transmitted through the germ line to produce progeny of the chimeric mouse.
The culturing of EC cells led to the isolation of embryonic stem (ES) cells. The optimal stage of embryonic development was determined by comparing cell-surface antigens and patterns of protein synthesis between EC cells and mouse embryonic cells at various stages (Evans and Kaufman 1981). In addition, the growth conditions used in maintaining EC cells were instrumental for the eventual isolation of ES cells. The transformed feeder cells used in some EC cell cultures, for example, were helpful in preventing differentiation of the embryonic cells. Evans and Kaufman (1981) were the first to isolate ES cells by explanting whole blastocysts. Independently, G.R. Martin (1981) was able to obtain ES cells by isolating the inner cell mass with immunosurgery. The isolated stem cells could propagate in culture and were able to differentiate into various cell types under specified growth environments. Unlike EC cells, the stem cell cultures were karyotypically normal. Bradley et al. (1984) showed that, when injected into a blastocyst, these stem cells could populate a chimeric mouse, further proving the pluripotency of the ES cells. Of major significance, though, was the ability to achieve germline transmission with the newly isolated cell cultures, a finding that could be confirmed through transmission of coat color. The cytokine leukemia inhibition factor (LIF) was later added to cultures to suppress differentiation. Before being cloned and characterized, LIF was initially added to EC cultures under the name DIA for differentiation inhibitory activity. With improvements in culturing conditions, ES cells from mouse strains other than 129 could be isolated.
As techniques were perfected in applying homologous recombination for gene targeting, researchers quickly realized that murine stem cells could be used as a means in which to make genetically altered mice. With the convergence of these technologies, a targeting construct is first delivered into cultured ES cells to introduce a desired mutation into a selected gene. The designated mutation is inserted into the targeted gene via recombination at sequences of genetic homology. The stem cell clone with the desired recombination event is then used to derive a mouse with the targeted mutation through injection into a blastocyst. The combination of homologous recombination and ES cells was first applied in gene targeting of hypoxanthine-guanosine phosphoribosyl transferase gene (HPRT). The HPRT gene was an ideal candidate to test gene targeting since both correction (Doetschman et al. 1987) and inactivation (Thomas and Capecchi 1987) of this gene could be easily tested in tissue culture. Later refinements allowed for the targeted inactivation of nonselectable genes such as int-2 and c-abl knockout mice (Mansour et al. 1988; Schwartzberg et al. 1989). These refinements included positive and negative drug selection (Mansour et al. 1988). Soon, the methods used to make knockout mice were modified to create other gene-targeted mutations. In this manner, conditional knockout mice, knock-in mice, and mice with subtle genetic mutations were generated. The significance of gene targeting was seen when Mario Capecchi, Oliver Smithies, and Martin Evans won the 2007 Nobel Prize in Physiology or Medicine. Additionally, the explosive growth in the number of animal models derived through gene targeting attests to the usefulness of these discoveries. With targeted gene inactivation in mice, the precise role of a gene can be discerned by analyzing its absence in vivo. Through gene targeting to disrupt genes or create subtle mutations in mice, human genetic disease could then be mimicked in an animal model. Therefore, using stem cells in combination with gene targeting has been a valuable means to gain insights into mammalian biology.
References
- Baharvand H, Matthaei KI. Culture condition difference for establishment of new embryonic stem cell lines from the C57BL/6 and BALB/c mouse strains. In Vitro Cell Dev Biol Anim. 2004;40:76–81. doi: 10.1290/1543-706x(2004)040<0076:ccdfeo>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines . Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- Brinster RL. The effect of cells transferred into the mouse blastocyst on subsequent development. J Exp Med. 1974;140:1049–56. doi: 10.1084/jem.140.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- Cole RJ, Paul J. Preimplantation Stages of Pregnancy. In: Wolstenholme GEW, O'Conner M, editors. Ciba Found Symp. Vol. 82. Little, Brown and Co; Boston: 1965. [Google Scholar]
- Dinkel A, Aicher WK, Warnatz K, Bürki K, Eibel H, Ledermann B. Efficient generation of transgenic BALB/c mice using BALB/c embryonic stem cells. J Immunol Methods. 1999;223:255–60. doi: 10.1016/s0022-1759(99)00005-8. [DOI] [PubMed] [Google Scholar]
- Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, Smithies O. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- Downing GJ, Battey JF. Technical assessment of the first 20 years of research using mouse embryonic stem cell lines. Stem Cells. 2004;22:1168–80. doi: 10.1634/stemcells.2004-0101. [DOI] [PubMed] [Google Scholar]
- Eakin GS, Hadjantonakis AK. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos . Nat Protoc. 2006;1:1145–53. doi: 10.1038/nprot.2006.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos . Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Mouse chimeras obtained by the injection of cells into the blastocyst. Nature. 1968;220:596–7. doi: 10.1038/220596a0. [DOI] [PubMed] [Google Scholar]
- Gossler A, Doetschman T, Korn R, Serfling E, Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci. 1986;83:9065–9. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–5. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- Köntgen F, Stewart CL. Simple screening procedure to detect gene targeting events in embryonic stem cells. Methods Enzymol. 1993;225:878–90. doi: 10.1016/0076-6879(93)25055-7. [DOI] [PubMed] [Google Scholar]
- Kosaka Y, Kobayashi N, Fukazawa T, Totsugawa T, Maruyama M, Yong C, Arata T, Ikeda H, Kobayashi K, Ueda T, Kurabayashi Y, Tanaka N. Lentivirus-based gene delivery in mouse embryonic stem cells. Artif Organs. 2004;28 :271–7. doi: 10.1111/j.1525-1594.2004.47297.x. [DOI] [PubMed] [Google Scholar]
- Ledermann B, Bürki K. Establishment of a germ-line competent C57BL/6 embryonic stem cell line. Exp Cell Res. 1991;197:254–8. doi: 10.1016/0014-4827(91)90430-3. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes . Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Martin GR, Evans MJ. The morphology and growth of a pluripotent teratocarcinoma cell line and its derivatives in tissue culture. Cell. 1974;2:163–72. doi: 10.1016/0092-8674(74)90090-7. [DOI] [PubMed] [Google Scholar]
- Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro . Proc Natl Acad Sci. 1975;72:1441–5. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma. Proc Natl Acad Sci. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Mortensen RM, Conner DA, Chao S, Geister-Lowrance AA, Seidman JG. Production of homozygous mutant ES cells with a single targeting construct. Mol C ell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Rossant J. Production and analysis of ES cell aggregation chimeras. In: Joyner AL, editor. Gene Targeting: a Practical Approach. 2. Oxford University Press; New York: 2000. pp. 177–206. [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–6. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Noben-Trauth N, Köhler G, Bürki K, Ledermann B. Efficient targeting of the IL-4 gene in a BALB/c embryonic stem cell line. Transgenic Res. 1996;5:487–91. doi: 10.1007/BF01980214. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germ line-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, McBurney MW, Gardner RL, Evans MJ. Fate of teratocarcinoma cells injected into early mouse embryos. Nature. 1975;258:70–73. doi: 10.1038/258070a0. [DOI] [PubMed] [Google Scholar]
- Pease S, Braghetta P, Gearing D, Grail D, Williams RL. Isolation of embryonic stem (ES) cells in media supplemented with recombinant leukemia inhibitory factor (LIF) Dev Biol. 1990;141:344–52. doi: 10.1016/0012-1606(90)90390-5. [DOI] [PubMed] [Google Scholar]
- Robertson E, Bradley A, Kuehn M, Evans M. Germ-line transmission of genes introduced into cultured pluripotential cells by retroviral vector. Nature. 1986;323:445–8. doi: 10.1038/323445a0. [DOI] [PubMed] [Google Scholar]
- Robertson EJ. Embryo-derived stem cells. In: Robertson EJ, editor. Teratocarcinomas and embryonic stem cells a practical approach. IRL Press; Oxford: 1987. pp. 71–112. [Google Scholar]
- Schoonjans L, Kreemers V, Danloy S, Moreadith RW, Laroche Y, Collen D. Improved generation of germ line-competent embryonic stem cell lines from inbred mouse strains. Stem Cells. 2003;21:90–7. doi: 10.1634/stemcells.21-1-90. [DOI] [PubMed] [Google Scholar]
- Schwartzberg PL, Goff SP, Robertson EJ. Germ-line transmission of a c-abl mutation produced by targeted gene disruption in ES cells . Science. 1989;246:799–803. doi: 10.1126/science.2554496. [DOI] [PubMed] [Google Scholar]
- Seong E, Saunders TL, Stewart CL, Burmeister M. To knockout in 129 or in C57BL/6: that is the question. Trends Genet. 2004;20:59–62. doi: 10.1016/j.tig.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Sherman MI. The culture of cells derived from mouse blastocysts. Cell. 1975;5:343–9. doi: 10.1016/0092-8674(75)90052-5. [DOI] [PubMed] [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Smith-Arica JR, Thomson AJ, Ansell R, Chiorini J, Davidson B, McWhir J. Infection efficiency of human and mouse embryonic stem cells using adenoviral and adeno-associated viral vectors. Cloning Stem Cells. 2003;5:51–62. doi: 10.1089/153623003321512166. [DOI] [PubMed] [Google Scholar]
- Solter D, Skreb N, Damjanov I. Extrauterine growth of mouse egg-cylinders results in malignant teratoma. Nature. 1970;227:503–4. doi: 10.1038/227503a0. [DOI] [PubMed] [Google Scholar]
- Solter D, Shevinsky L, Knowles BB, Strickland S. The induction of antigenic changes in a teratocarcinoma stem cell line (F9) by retinoic acid. Dev Biol. 1979;70:515–21. doi: 10.1016/0012-1606(79)90043-5. [DOI] [PubMed] [Google Scholar]
- Stanford WL, Cohn JB, Cordes SP. Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–68. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- Stevens LC, Little CC. Spontaneous Testicular Teratomas in an Inbred Strain of Mice. Proc Natl Acad Sci. 1954;40:1080–7. doi: 10.1073/pnas.40.11.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens LC. The development of transplantable teratocarcinomas from intratesticular grafts of pre- and postimplantation mouse embryos. Dev Biol. 1970;21:364–82. doi: 10.1016/0012-1606(70)90130-2. [DOI] [PubMed] [Google Scholar]
- Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells . Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination . Proc Natl Acad Sci. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–7. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]