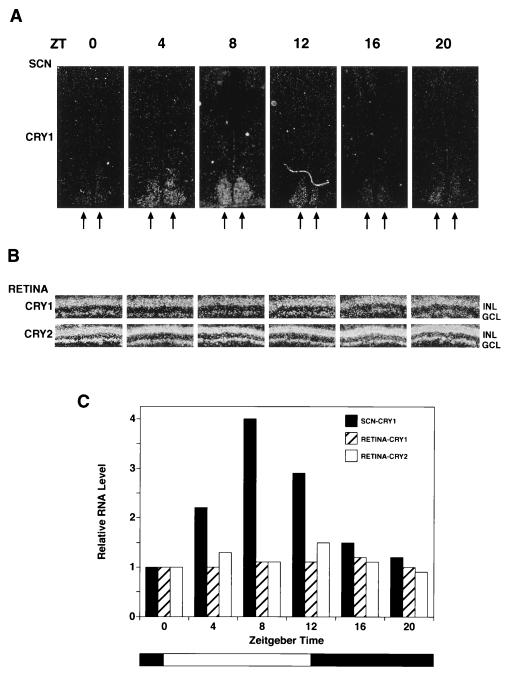
Figure 4.
Circadian expression of Cry1 and Cry2 mRNA in the mouse SCN and retina. Mice kept under a 12-hr light/12-he dark cycle were sacrificed every 4 hr, and retinal sections and coronal brain sections encompassing the SCN were prepared for in situ hybridization using 35S-labeled mCry1 and mCry2 RNA probes. Only the results of mCry1 hybridization to SCN are shown because the SCN signal with mCry2 was very weak at all Zeitgeber time (ZT) points and not amenable to quantitative analysis. (A) Expression of mCry1 in SCN. In situ hybridization of mCry1 with coronal brain sections passing through the SCN region are shown. Seven sections (20 μm) were made for each point and the one with the strongest signal for each point are shown in these micrograms. The double arrows indicate the SCN. (B) Expression of mCry1 and mCry2 in the retina. Retinal sections from the central part of the retina were hybridized with mCry1 and mCry2. The GCL and the INL are indicated. (C) Quantitative analyses of Cryptochrome expression in the SCN and the retina as a function of light/dark cycles (34). Grain density from autoradiograms of all section containing SCN were measured and the total grain density for each time point is expressed relative to zeitgeber time (ZT) = 0 value, which represents the values just before turning on the light (at 0800 hr). For retina, for each time point, the grain density was measured in the GCL and INL of the entire retinal region present in the autoradiogram and normalized for the length of the retina present in each section. The intensity for mCry1 and mCry2 are expressed relative to the values of mCry1 and mCry2 at ZT = 0, respectively. In absolute terms, the grain density with mCry2 probe was about 3-fold higher than that obtained with the mCry1 probe at all times.