FIGURE 2.
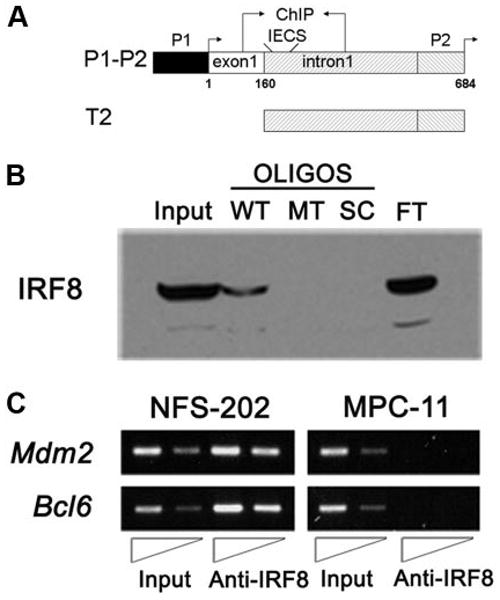
IRF8 binds sequences from the Mdm2 5′ regulatory region in vitro and in vivo. A, Schematic drawing of the Mdm2 promoter and reporter constructs P1–P2 and T2. Shown are the locations of the basal (P1) and p53-responsive promoters (P2) with their associated transcription start sites (arrows) and exon 1. The location of an IECS-like element and the region examined in ChIP analyses are also shown. The start site of exon 1 is indicated as +1. B, Western blot analyses of oligonucleotide pull-downs using the wild-type IECS-like (WT; core -GAAAAGAGGGAA-, italic denotes the two core elements), mutant (MT; core -GCAGAGAGTGTG-, underscore denotes the mutation), and scrambled (SC; core -GGAGAG GAGCGG-) oligonucleotides (OLIGOS). Also shown are control lanes with 10% of input nuclear protein (Input) and flow-through unbound protein (FT). C, ChIP analysis for in vivo binding of IRF8 to mouse Mdm2 P2 promoter. Anti-IRF8 Ab was used to precipitate fragments of genomic DNA from NFS-202 cells. The binding of IRF8 to Mdm2 P2 promoter is shown, together with its binding to Bcl6 promoter as a positive control. The same experiments were performed using DNA from MPC-11 cells that express little if any IRF8.
